Genomic insights into plant growth promoting rhizobia capable of enhancing soybean germination under drought stress
- PMID: 31296165
- PMCID: PMC6624879
- DOI: 10.1186/s12866-019-1536-1
Genomic insights into plant growth promoting rhizobia capable of enhancing soybean germination under drought stress
Abstract
Background: The role of soil microorganisms in plant growth, nutrient utilization, drought tolerance as well as biocontrol activity cannot be over-emphasized, especially in this era when food crisis is a global challenge. This research was therefore designed to gain genomic insights into plant growth promoting (PGP) Rhizobium species capable of enhancing soybean (Glycine max L.) seeds germination under drought condition.
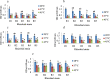
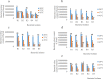
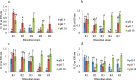
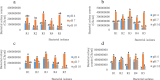
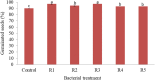
Results: Rhizobium sp. strain R1, Rhizobium tropici strain R2, Rhizobium cellulosilyticum strain R3, Rhizobium taibaishanense strain R4 and Ensifer meliloti strain R5 were found to possess the entire PGP traits tested. Specifically, these rhizobial strains were able to solubilize phosphate, produce exopolysaccharide (EPS), 1-aminocyclopropane-1-carboxylate (ACC), siderophore and indole-acetic-acid (IAA). These strains also survived and grew at a temperature of 45 °C and in an acidic condition with a pH 4. Consequently, all the Rhizobium strains enhanced the germination of soybean seeds (PAN 1532 R) under drought condition imposed by 4% poly-ethylene glycol (PEG); nevertheless, Rhizobium sp. strain R1 and R. cellulosilyticum strain R3 inoculations were able to improve seeds germination more than R2, R4 and R5 strains. Thus, genomic insights into Rhizobium sp. strain R1 and R. cellulosilyticum strain R3 revealed the presence of some genes with their respective proteins involved in symbiotic establishment, nitrogen fixation, drought tolerance and plant growth promotion. In particular, exoX, htrA, Nif, nodA, eptA, IAA and siderophore-producing genes were found in the two rhizobial strains.
Conclusions: Therefore, the availability of the whole genome sequences of R1 and R3 strains may further be exploited to comprehend the interaction of drought tolerant rhizobia with soybean and other legumes and the PGP ability of these rhizobial strains can also be harnessed for biotechnological application in the field especially in semiarid and arid regions of the globe.
Keywords: Drought stress; Nitrogen fixation; PGP; Soybean; Symbiotic establishment; Whole genome sequences.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Effects of rhizobia and arbuscular mycorrhizal fungi on yield, size distribution and fatty acid of soybean seeds grown under drought stress.Microbiol Res. 2021 Jan;242:126640. doi: 10.1016/j.micres.2020.126640. Epub 2020 Nov 2. Microbiol Res. 2021. PMID: 33223380
-
Physiological and genomic insights into a psychrotrophic drought-tolerant bacterial consortium for crop improvement in cold, semiarid regions.Microbiol Res. 2024 Sep;286:127818. doi: 10.1016/j.micres.2024.127818. Epub 2024 Jun 26. Microbiol Res. 2024. PMID: 38970906
-
Genomic characterization of Ensifer aridi, a proposed new species of nitrogen-fixing rhizobium recovered from Asian, African and American deserts.BMC Genomics. 2017 Jan 14;18(1):85. doi: 10.1186/s12864-016-3447-y. BMC Genomics. 2017. PMID: 28088165 Free PMC article.
-
Challenges to rhizobial adaptability in a changing climate: Genetic engineering solutions for stress tolerance.Microbiol Res. 2024 Nov;288:127886. doi: 10.1016/j.micres.2024.127886. Epub 2024 Aug 31. Microbiol Res. 2024. PMID: 39232483 Review.
-
Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean.J Biotechnol. 2011 Aug 20;155(1):11-9. doi: 10.1016/j.jbiotec.2011.03.016. Epub 2011 Mar 30. J Biotechnol. 2011. PMID: 21458507 Review.
Cited by
-
Microbial Inoculants as Plant Biostimulants: A Review on Risk Status.Life (Basel). 2022 Dec 21;13(1):12. doi: 10.3390/life13010012. Life (Basel). 2022. PMID: 36675961 Free PMC article. Review.
-
Diversity and function of soybean rhizosphere microbiome under nature farming.Front Microbiol. 2023 Mar 1;14:1130969. doi: 10.3389/fmicb.2023.1130969. eCollection 2023. Front Microbiol. 2023. PMID: 36937301 Free PMC article.
-
The Application of Arbuscular Mycorrhizal Fungi as Microbial Biostimulant, Sustainable Approaches in Modern Agriculture.Plants (Basel). 2023 Aug 29;12(17):3101. doi: 10.3390/plants12173101. Plants (Basel). 2023. PMID: 37687348 Free PMC article. Review.
-
Bacterial Exopolysaccharides: Insight into Their Role in Plant Abiotic Stress Tolerance.J Microbiol Biotechnol. 2021 Aug 28;31(8):1045-1059. doi: 10.4014/jmb.2105.05009. J Microbiol Biotechnol. 2021. PMID: 34226402 Free PMC article. Review.
-
Exogenous Substances Used to Relieve Plants from Drought Stress and Their Associated Underlying Mechanisms.Int J Mol Sci. 2024 Aug 26;25(17):9249. doi: 10.3390/ijms25179249. Int J Mol Sci. 2024. PMID: 39273198 Free PMC article. Review.
References
-
- Ormeño-Orrillo E, Menna P, Almeida LGP, Ollero FJ, Nicolás MF, Rodrigues EP, et al. Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.) BMC Genomics. 2012;13:735. doi: 10.1186/1471-2164-13-735. - DOI - PMC - PubMed
-
- Grover M, Ali SZ, Sandhya V, Rasul A, Venkateswarlu B. Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J Microbiol Biotechnol. 2011;27:1231–1240. doi: 10.1007/s11274-010-0572-7. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

