Integrated transcriptome, small RNA, and degradome analysis reveals the complex network regulating starch biosynthesis in maize
- PMID: 31296166
- PMCID: PMC6625009
- DOI: 10.1186/s12864-019-5945-1
Integrated transcriptome, small RNA, and degradome analysis reveals the complex network regulating starch biosynthesis in maize
Abstract
Background: Starch biosynthesis in endosperm is a key process influencing grain yield and quality in maize. Although a number of starch biosynthetic genes have been well characterized, the mechanisms by which the expression of these genes is regulated, especially in regard to microRNAs (miRNAs), remain largely unclear.
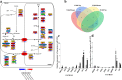
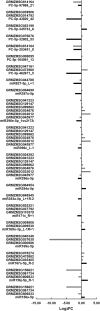
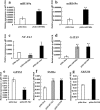
Results: Sequence data for small RNAs, degradome, and transcriptome of maize endosperm at 15 and 25 d after pollination (DAP) from inbred lines Mo17 and Ji419, which exhibit distinct starch content and starch granule structure, revealed the mediation of starch biosynthetic pathways by miRNAs. Transcriptome analysis of these two lines indicated that 33 of 40 starch biosynthetic genes were differentially expressed, of which 12 were up-regulated in Ji419 at 15 DAP, one was up-regulated in Ji419 at 25 DAP, 14 were up-regulated in Ji419 at both 15 and 25 DAP, one was down-regulated in Ji419 at 15 DAP, two were down-regulated in Ji419 at 25 DAP, and three were up-regulated in Ji419 at 15 DAP and down-regulated in Ji419 at 25 DAP, compared with Mo17. Through combined analyses of small RNA and degradome sequences, 22 differentially expressed miRNAs were identified, including 14 known and eight previously unknown miRNAs that could target 35 genes. Furthermore, a complex co-expression regulatory network was constructed, in which 19 miRNAs could modulate starch biosynthesis in endosperm by tuning the expression of 19 target genes. Moreover, the potential operation of four miRNA-mediated pathways involving transcription factors, miR169a-NF-YA1-GBSSI/SSIIIa and miR169o-GATA9-SSIIIa/SBEIIb, was validated via analyses of expression pattern, transient transformation assays, and transactivation assays.
Conclusion: Our results suggest that miRNAs play a critical role in starch biosynthesis in endosperm, and that miRNA-mediated networks could modulate starch biosynthesis in this tissue. These results have provided important insights into the molecular mechanism of starch biosynthesis in developing maize endosperm.
Keywords: Endosperm; Maize; Regulatory network; Starch biosynthesis; miRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

