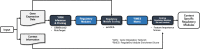
CONFIGURE: A pipeline for identifying context specific regulatory modules from gene expression data and its application to breast cancer
- PMID: 31296219
- PMCID: PMC6624175
- DOI: 10.1186/s12920-019-0515-6
CONFIGURE: A pipeline for identifying context specific regulatory modules from gene expression data and its application to breast cancer
Abstract
Background: Gene expression data is widely used for identifying subtypes of diseases such as cancer. Differentially expressed gene analysis and gene set enrichment analysis are widely used for identifying biological mechanisms at the gene level and gene set level, respectively. However, the results of differentially expressed gene analysis are difficult to interpret and gene set enrichment analysis does not consider the interactions among genes in a gene set.
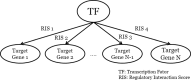
Results: We present CONFIGURE, a pipeline that identifies context specific regulatory modules from gene expression data. First, CONFIGURE takes gene expression data and context label information as inputs and constructs regulatory modules. Then, CONFIGURE makes a regulatory module enrichment score (RMES) matrix of enrichment scores of the regulatory modules on samples using the single-sample GSEA method. CONFIGURE calculates the importance scores of the regulatory modules on each context to rank the regulatory modules. We evaluated CONFIGURE on the Cancer Genome Atlas (TCGA) breast cancer RNA-seq dataset to determine whether it can produce biologically meaningful regulatory modules for breast cancer subtypes. We first evaluated whether RMESs are useful for differentiating breast cancer subtypes using a multi-class classifier and one-vs-rest binary SVM classifiers. The multi-class and one-vs-rest binary classifiers were trained using the RMESs as features and outperformed baseline classifiers. Furthermore, we conducted literature surveys on the basal-like type specific regulatory modules obtained by CONFIGURE and showed that highly ranked modules were associated with the phenotypes of basal-like type breast cancers.
Conclusions: We showed that enrichment scores of regulatory modules are useful for differentiating breast cancer subtypes and validated the basal-like type specific regulatory modules by literature surveys. In doing so, we found regulatory module candidates that have not been reported in previous literature. This demonstrates that CONFIGURE can be used to predict novel regulatory markers which can be validated by downstream wet lab experiments. We validated CONFIGURE on the breast cancer RNA-seq dataset in this work but CONFIGURE can be applied to any gene expression dataset containing context information.
Keywords: Breast cancer subtype; Context specific regulatory module; Feature importance score; Gene regulatory network inference; Single sample GSEA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Identification of breast cancer prognostic modules via differential module selection based on weighted gene Co-expression network analysis.Biosystems. 2021 Jan;199:104317. doi: 10.1016/j.biosystems.2020.104317. Epub 2020 Dec 3. Biosystems. 2021. PMID: 33279569
-
RNA-Seq-Based Breast Cancer Subtypes Classification Using Machine Learning Approaches.Comput Intell Neurosci. 2020 Oct 29;2020:4737969. doi: 10.1155/2020/4737969. eCollection 2020. Comput Intell Neurosci. 2020. PMID: 33178256 Free PMC article.
-
Comparisons of gene coexpression network modules in breast cancer and ovarian cancer.BMC Syst Biol. 2018 Apr 11;12(Suppl 1):8. doi: 10.1186/s12918-018-0530-9. BMC Syst Biol. 2018. PMID: 29671401 Free PMC article.
-
Evaluation of gene-drug common module identification methods using pharmacogenomics data.Brief Bioinform. 2021 May 20;22(3):bbaa087. doi: 10.1093/bib/bbaa087. Brief Bioinform. 2021. PMID: 32591780 Review.
-
Comparing enrichment analysis and machine learning for identifying gene properties that discriminate between gene classes.Brief Bioinform. 2020 May 21;21(3):803-814. doi: 10.1093/bib/bbz028. Brief Bioinform. 2020. PMID: 30895300 Review.
Cited by
-
The Atlas of Inflammation Resolution (AIR).Mol Aspects Med. 2020 Aug;74:100894. doi: 10.1016/j.mam.2020.100894. Epub 2020 Sep 3. Mol Aspects Med. 2020. PMID: 32893032 Free PMC article. Review.
References
-
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in pdgfra, idh1, egfr, and nf1. Cancer cell. 2010;17(1):98–110. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous