Cell-free DNA analysis in healthy individuals by next-generation sequencing: a proof of concept and technical validation study
- PMID: 31296838
- PMCID: PMC6624284
- DOI: 10.1038/s41419-019-1770-3
Cell-free DNA analysis in healthy individuals by next-generation sequencing: a proof of concept and technical validation study
Abstract
Pre-symptomatic screening of genetic alterations might help identify subpopulations of individuals that could enter into early access prevention programs. Since liquid biopsy is minimally invasive it can be used for longitudinal studies in healthy volunteers to monitor events of progression from normal tissue to pre-cancerous and cancerous condition. Yet, cell-free DNA (cfDNA) analysis in healthy individuals comes with substantial challenges such as the lack of large cohort studies addressing the impact of mutations in healthy individuals or the low abundance of cfDNA in plasma. In this study, we aimed to investigate the technical feasibility of cfDNA analysis in a collection of 114 clinically healthy individuals. We first addressed the impact of pre-analytical factors such as cfDNA yield and quality on sequencing performance and compared healthy to cancer donor samples. We then confirmed the validity of our testing strategy by evaluating the mutational status concordance in matched tissue and plasma specimens collected from cancer patients. Finally, we screened our group of healthy donors for genetic alterations, comparing individuals who did not develop any tumor to patients who developed either a benign neoplasm or cancer during 1-10 years of follow-up time. To conclude, we have established a rapid and reliable liquid biopsy workflow that allowed us to study genomic alterations with a limit of detection as low as 0.08% of variant allelic frequency in healthy individuals. We detected pathogenic cancer mutations in four healthy donors that later developed a benign neoplasm or invasive breast cancer up to 10 years after blood collection. Even though larger prospective studies are needed to address the specificity and sensitivity of liquid biopsy as a clinical tool for early cancer detection, systematic screening of healthy individuals will help understanding early events of tumor formation.
Conflict of interest statement
P.J., L.Q., N.A. and M.S. were at the time of this project establishment part of the Scientific Advisory Board of the Bioscience Institute. I.A. and P.J. report grants from BMS and non-financial support from Thermo Fisher Scientific. G.M. is the Founder and CEO of the Bioscience Institute SpA. L.Q. no longer serves on the Bioscience Institute Scientific Board and currently is an Employer of Thermo Fisher Scientific. He has joined Thermo Fisher Scientific when this project was already accomplished and only manuscript had to be written. L.B. reports grants and personal fees from Roche, grants and personal fees from MSD, personal fees from BMS, personal fees from Astra Zeneca. N.A. is a paid consultant for pharmaceutical and insurance companies with an interest in liquid biopsy, and he is listed as inventor in several patent applications related to cancer detection and treatment. M.S. has received research funds from Puma Biotechnology, Daiichi-Sankio, Immunomedics, Targimmune and Menarini Ricerche, and is a cofounder of Medendi Medical Travel. The remaining authors declare that they have no conflict of interest.
Figures
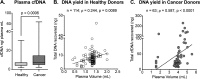
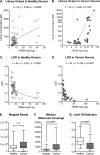
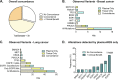

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

