Novel Protein Mg2046 Regulates Magnetosome Synthesis in Magnetospirillum gryphiswaldense MSR-1 by Modulating a Proper Redox Status
- PMID: 31297108
- PMCID: PMC6607277
- DOI: 10.3389/fmicb.2019.01478
Novel Protein Mg2046 Regulates Magnetosome Synthesis in Magnetospirillum gryphiswaldense MSR-1 by Modulating a Proper Redox Status
Abstract
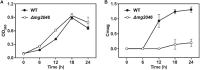
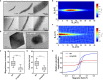
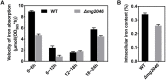
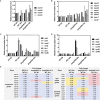
Magnetotactic bacteria (MTB) are a large, polyphyletic group of aquatic microorganisms capable of absorbing large amounts of iron and synthesizing intercellular nano-scaled nanoparticles termed magnetosomes. In our previous transcriptomic studies, we discovered that a novel gene (MGMSRv2_2046, termed as mg2046) in Magnetospirillum gryphiswaldense strain MSR-1 was significantly up-regulated during the period of magnetosome synthesis. In the present study, we constructed a MSR-1 mutant strain with deletion of mg2046 (termed Δmg2046) in order to evaluate the role of this gene in cell physiological status and magnetosome formation process. In comparison with wild-type MSR-1, Δmg2046 showed similar cell growth, but much lower cell magnetic response, smaller number and size of magnetosomes, and reduced iron absorption ability. mg2046 deletion evidently disrupted iron uptake, and redox equilibrium, and strongly inhibited transcription of dissimilatory denitrification pathway genes. Our experimental findings, taken together with results of gene homology analysis, indicate that Mg2046 acts as a positive regulator in MSR-1 under microaerobic conditions, responding to hypoxia signals and participating in regulation of oxygen metabolism, in part as a co-regulator of dissimilatory denitrification pathway with oxygen sensor MgFnr (MGMSRv2_2946, termed as Mg2946). Mg2046 is clearly involved in coupled regulation of cellular oxygen, iron and nitrogen metabolism under micro-aerobic or anaerobic conditions. Our findings help explain how MSR-1 cells initiate dissimilatory denitrification pathway and overcome energy deficiency under microaerobic conditions, and have broader implications regarding bacterial survival and energy metabolism strategies under hypoxia.
Keywords: Magnetospirillum gryphiswaldense; Mg2046; dissimilatory denitrification pathway; magnetosome; redox status; regulator.
Figures


Similar articles
-
The oxygen sensor MgFnr controls magnetite biomineralization by regulation of denitrification in Magnetospirillum gryphiswaldense.BMC Microbiol. 2014 Jun 10;14:153. doi: 10.1186/1471-2180-14-153. BMC Microbiol. 2014. PMID: 24915802 Free PMC article.
-
Iron response regulator protein IrrB in Magnetospirillum gryphiswaldense MSR-1 helps control the iron/oxygen balance, oxidative stress tolerance, and magnetosome formation.Appl Environ Microbiol. 2015 Dec;81(23):8044-53. doi: 10.1128/AEM.02585-15. Epub 2015 Sep 18. Appl Environ Microbiol. 2015. PMID: 26386052 Free PMC article.
-
Transcriptome analysis reveals physiological characteristics required for magnetosome formation in Magnetospirillum gryphiswaldense MSR-1.Environ Microbiol Rep. 2016 Jun;8(3):371-81. doi: 10.1111/1758-2229.12395. Epub 2016 May 16. Environ Microbiol Rep. 2016. PMID: 27043321
-
Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense.Arch Microbiol. 2004 Jan;181(1):1-7. doi: 10.1007/s00203-003-0631-7. Epub 2003 Dec 11. Arch Microbiol. 2004. PMID: 14668979 Review.
-
The bacterial magnetosome: a unique prokaryotic organelle.J Mol Microbiol Biotechnol. 2013;23(1-2):63-80. doi: 10.1159/000346543. Epub 2013 Apr 18. J Mol Microbiol Biotechnol. 2013. PMID: 23615196 Review.
Cited by
-
Nitric oxide sensor NsrR is the key direct regulator of magnetosome formation and nitrogen metabolism in Magnetospirillum.Nucleic Acids Res. 2024 Apr 12;52(6):2924-2941. doi: 10.1093/nar/gkad1230. Nucleic Acids Res. 2024. PMID: 38197240 Free PMC article.
-
Key Signatures of Magnetofossils Elucidated by Mutant Magnetotactic Bacteria and Micromagnetic Calculations.J Geophys Res Solid Earth. 2022 Jan;127(1):e2021JB023239. doi: 10.1029/2021jb023239. Epub 2021 Dec 28. J Geophys Res Solid Earth. 2022. PMID: 35444924 Free PMC article.
-
Metabolic characterisation of Magnetospirillum gryphiswaldense MSR-1 using LC-MS-based metabolite profiling.RSC Adv. 2020 Sep 2;10(54):32548-32560. doi: 10.1039/d0ra05326k. eCollection 2020 Sep 1. RSC Adv. 2020. PMID: 35516490 Free PMC article.
-
Fusion expression of nanobodies specific for the insecticide fipronil on magnetosomes in Magnetospirillum gryphiswaldense MSR-1.J Nanobiotechnology. 2021 Jan 19;19(1):27. doi: 10.1186/s12951-021-00773-z. J Nanobiotechnology. 2021. PMID: 33468141 Free PMC article.
References
-
- Baumgartner J., Morin G., Menguy N., Perez Gonzalez T., Widdrat M., Cosmidis J., et al. (2013). Magnetotactic bacteria form magnetite from a phosphate-rich ferric hydroxide via nanometric ferric (oxyhydr) oxide intermediates. Proc. Natl. Acad. Sci. U.S.A. 110 14883–14888. 10.1073/pnas.1307119110 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

