Quality Controls in Ligand Binding Assays: Recommendations and Best Practices for Preparation, Qualification, Maintenance of Lot to Lot Consistency, and Prevention of Assay Drift
- PMID: 31297703
- PMCID: PMC6647453
- DOI: 10.1208/s12248-019-0354-6
Quality Controls in Ligand Binding Assays: Recommendations and Best Practices for Preparation, Qualification, Maintenance of Lot to Lot Consistency, and Prevention of Assay Drift
Abstract
Quality controls (QCs) are the primary indices of assay performance and an important tool in assay lifecycle management. Inclusion of QCs in the testing process allows for the detection of system errors and ongoing assessment of the reliability of the assay. Changes in the performance of QCs are indicative of changes in the assay behavior caused by unintended alterations to reagents or to the operating conditions. The focus of this publication is management of QC life cycle. A consensus view of the ligand binding assay (LBA) community on the best practices for factors that are critical to QC life cycle management including QC preparation, qualification, and trending is presented here.
Keywords: LBA; life cycle management; qualification; quality control; trending.
Figures

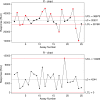

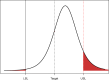


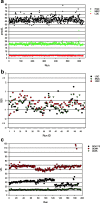
References
-
- DeSilva B, Smith W, Weiner R, Kelley M, Smolec J, Lee B, et al. Recommendations for the bioanalytical method validation of ligand-binding assays to support pharmacokinetic assessments of macromolecules. AAPS J. 2003;20(11):1885–1900. - PubMed
-
- US Food and Drug Administration . Guidance for industry: bioanalytical method validation. Silver Spring: Center for Drug Evaluation and Research; 2018.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

