Prediction of interresidue contacts with DeepMetaPSICOV in CASP13
- PMID: 31298436
- PMCID: PMC6899903
- DOI: 10.1002/prot.25779
Prediction of interresidue contacts with DeepMetaPSICOV in CASP13
Abstract
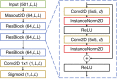
In this article, we describe our efforts in contact prediction in the CASP13 experiment. We employed a new deep learning-based contact prediction tool, DeepMetaPSICOV (or DMP for short), together with new methods and data sources for alignment generation. DMP evolved from MetaPSICOV and DeepCov and combines the input feature sets used by these methods as input to a deep, fully convolutional residual neural network. We also improved our method for multiple sequence alignment generation and included metagenomic sequences in the search. We discuss successes and failures of our approach and identify areas where further improvements may be possible. DMP is freely available at: https://github.com/psipred/DeepMetaPSICOV.
Keywords: deep learning; machine learning; metagenomics; neural networks; protein contact prediction; protein structure prediction.
© 2019 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals, Inc.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

