Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil
- PMID: 31299891
- PMCID: PMC6626388
- DOI: 10.1186/s12866-019-1531-6
Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil
Abstract
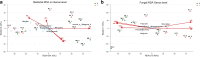
Background: Fusarium wilt of banana (Musa spp.) caused by the fungal pathogen Fusarium oxysporum f. sp. cubense (Foc) is a typical soilborne disease, that severely devastates the banana industry worldwide, and soil microbial diversity is closely related to the spread of Fusarium wilt. To understand the relationship between microbial species and Fusarium wilt, it is important to understand the microbial diversity of the Fusarium wilt-diseased and disease-free soils from banana fields.
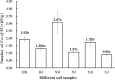
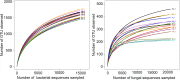
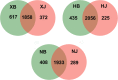
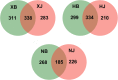
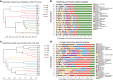
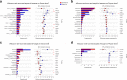
Results: Based on sequencing analysis of the bacterial 16S rRNA genes and fungal internal transcribed spacer (ITS) sequences, Foc abundance, fungal or bacterial richness and diversity were higher in the diseased soils than in the disease-free soils. Although Ascomycota and Zygomycota were the most abundant fungi phyla in all soil samples, Ascomycota abundance was significantly reduced in the disease-free soils. Mortierella (36.64%) was predominant in the disease-free soils. Regarding bacterial phyla, Proteobacteria, Acidobacteria, Chloroflexi, Firmicutes, Actinobacteria, Gemmatimonadetes, Bacteroidetes, Nitrospirae, Verrucomicrobia and Planctomycetes were dominant phyla in all soil samples. In particular, Firmicutes contributed 16.20% of the total abundance of disease-free soils. At the bacterial genus level, Bacillus, Lactococcus and Pseudomonas were abundant in disease-free soils with abundances of 8.20, 5.81 and 2.71%, respectively; lower abundances, of 4.12, 2.35 and 1.36%, respectively, were found in diseased soils. The distribution characteristics of fungal and bacterial genera may contribute to the abundance decrease of Foc in the disease-free soils.
Conclusion: Unique distributions of bacteria and fungi were observed in the diseased and disease-free soil samples from banana fields. These specific genera are useful for constructing a healthy microbial community structure of soil.
Keywords: Bacterial and fungal communities; Banana Fusarium wilt; Environmental variables; Pathogen abundance.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Bonanomi G, Antignani V, Capodilupo M, Scala F. Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biol Biochem. 2010;42:136–144. doi: 10.1016/j.soilbio.2009.10.012. - DOI
-
- Van Bruggen AHC, Sharma K, Kaku E, Karfopoulos S, Zelenev VV, Blok WJ. Soil health indicators and Fusarium wilt suppression in organically and conventionally managed green house soils. Appl Soil Ecol. 2015;86:192–201. doi: 10.1016/j.apsoil.2014.10.014. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

