Validation of 46 loci associated with female fertility traits in cattle
- PMID: 31299913
- PMCID: PMC6624949
- DOI: 10.1186/s12864-019-5935-3
Validation of 46 loci associated with female fertility traits in cattle
Abstract
Background: Subfertility is one challenge facing the dairy industry as the average Holstein heifer conception rate (HCR), the proportion of heifers that conceive and maintain a pregnancy per breeding, is estimated at 55-60%. Of the loci associated with HCR, few have been validated in an independent cattle population, limiting their usefulness for selection or furthering our understanding of the mechanisms involved in successful pregnancy. Therefore, the objectives here were to identify loci associated with HCR: 1) to the first artificial insemination (AI) service (HCR1), 2) to repeated AI services required for a heifer to conceive (TBRD) and 3) to validate loci previously associated with fertility. Breeding and health records from 3359 Holstein heifers were obtained after heifers were bred by AI at observed estrus, with pregnancy determined at day 35 via palpation. Heifer DNA was genotyped using the Illumina BovineHD BeadChip, and genome-wide association analyses (GWAA) were performed with additive, dominant and recessive models using the Efficient Mixed Model Association eXpedited (EMMAX) method with a relationship matrix for two phenotypes. The HCR1 GWAA compared heifers that were pregnant after the first AI service (n = 497) to heifers that were open following the first AI service (n = 405), which included those that never conceived. The TBRD GWAA compared only those heifers which did conceive, across variable numbers of AI service (n = 712). Comparison of loci previously associated with fertility, HCR1 or TBRD were considered the same locus for validation when in linkage disequilibrium (D' > 0.7).
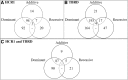
Results: The HCR1 GWAA identified 116, 187 and 28 loci associated (P < 5 × 10- 8) in additive, dominant and recessive models, respectively. The TBRD GWAA identified 235, 362, and 69 QTL associated (P < 5 × 10- 8) with additive, dominant and recessive models, respectively. Loci previously associated with fertility were in linkage disequilibrium with 22 loci shared with HCR1 and TBRD, 5 HCR1 and 19 TBRD loci.
Conclusions: Loci associated with HCR1 and TBRD that have been identified and validated can be used to improve HCR through genomic selection, and to better understand possible mechanisms associated with subfertility.
Keywords: Cattle; Conception rate; Dairy; Fertility; Loci.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Lucy MC. Reproductive loss in high-producing dairy cattle: where will it end? J Dairy Sci. 2001;84(6):1277–1293. - PubMed
-
- Beerda B, Wyszynka-Koko J, te Pas MFW, de Wit AAC, Veerkamp RF. Expression profiles of genes regulating dairy cow fertility: recent findings, ongoing activities and future possibilities. Animal. 2008;2(8):1158–1167. - PubMed
-
- Weigel KA, Rekaya R. Genetic parameters for reproductive traits of Holstein cattle in California and Minnesota. J Dairy Sci. 2000;83(5):1072–1080. - PubMed
-
- Andersen-Ranberg IM, Klemetsdal G, Heringstad B, Steine T. Heritabilities, genetic correlations, and genetic change for female fertility and protein yield in Norwegian dairy cattle. J Dairy Sci. 2005;88(1):348–355. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

