Best fitting tumor growth models of the von Bertalanffy-PütterType
- PMID: 31299926
- PMCID: PMC6624893
- DOI: 10.1186/s12885-019-5911-y
Best fitting tumor growth models of the von Bertalanffy-PütterType
Abstract
Background: Longitudinal studies of tumor volume have used certain named mathematical growth models. The Bertalanffy-Pütter differential equation unifies them: It uses five parameters, amongst them two exponents related to tumor metabolism and morphology. Each exponent-pair defines a unique three-parameter model of the Bertalanffy-Pütter type, and the above-mentioned named models correspond to specific exponent-pairs. Amongst these models we seek the best fitting one.
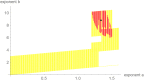
Method: The best fitting model curve within the Bertalanffy-Pütter class minimizes the sum of squared errors (SSE). We investigate also near-optimal model curves; their SSE is at most a certain percentage (e.g. 1%) larger than the minimal SSE. Models with near-optimal curves are visualized by the region of their near-optimal exponent pairs. While there is barely a visible difference concerning the goodness of fit between the best fitting and the near-optimal model curves, there are differences in the prognosis, whence the near-optimal models are used to assess the uncertainty of extrapolation.
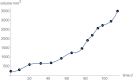
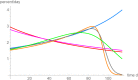
Results: For data about the growth of an untreated tumor we found the best fitting growth model which reduced SSE by about 30% compared to the hitherto best fit. In order to analyze the uncertainty of prognosis, we repeated the search for the optimal and near-optimal exponent-pairs for the initial segments of the data (meaning the subset of the data for the first n days) and compared the prognosis based on these models with the actual data (i.e. the data for the remaining days). The optimal exponent-pairs and the regions of near-optimal exponent-pairs depended on how many data-points were used. Further, the regions of near-optimal exponent-pairs were larger for the first initial segments, where fewer data were used.
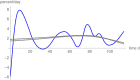
Conclusion: While for each near optimal exponent-pair its best fitting model curve remained close to the fitted data points, the prognosis using these model curves differed widely for the remaining data, whence e.g. the best fitting model for the first 65 days of growth was not capable to inform about tumor size for the remaining 49 days. For the present data, prognosis appeared to be feasible for a time span of ten days, at most.
Keywords: Bertalanffy-Pütter growth models; Cancer; Simulated annealing; Tumor growth.
Conflict of interest statement
The authors declare that they have no conceivable competing interests.
Figures






References
-
- Wheldon, T.E. Mathematical models in Cancer research, Bristol (UK): Adam Hilger1988.
-
- Michor F. Evolutionary dynamics of cancer. Doctoral thesis. Cambridge: Harvard Univ; 2005.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

