DNA Methylation Markers for Breast Cancer Detection in the Developing World
- PMID: 31300453
- PMCID: PMC6825533
- DOI: 10.1158/1078-0432.CCR-18-3277
DNA Methylation Markers for Breast Cancer Detection in the Developing World
Abstract
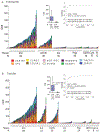
Purpose: An unmet need in low-resource countries is an automated breast cancer detection assay to prioritize women who should undergo core breast biopsy and pathologic review. Therefore, we sought to identify and validate a panel of methylated DNA markers to discriminate between cancer and benign breast lesions using cells obtained by fine-needle aspiration (FNA).Experimental Design: Two case-control studies were conducted comparing cancer and benign breast tissue identified from clinical repositories in the United States, China, and South Africa for marker selection/training (N = 226) and testing (N = 246). Twenty-five methylated markers were assayed by Quantitative Multiplex-Methylation-Specific PCR (QM-MSP) to select and test a cancer-specific panel. Next, a pilot study was conducted on archival FNAs (49 benign, 24 invasive) from women with mammographically suspicious lesions using a newly developed, 5-hour, quantitative, automated cartridge system. We calculated sensitivity, specificity, and area under the receiver-operating characteristic curve (AUC) compared with histopathology for the marker panel.
Results: In the discovery cohort, 10 of 25 markers were selected that were highly methylated in breast cancer compared with benign tissues by QM-MSP. In the independent test cohort, this panel yielded an AUC of 0.937 (95% CI = 0.900-0.970). In the FNA pilot, we achieved an AUC of 0.960 (95% CI = 0.883-1.0) using the automated cartridge system.
Conclusions: We developed and piloted a fast and accurate methylation marker-based automated cartridge system to detect breast cancer in FNA samples. This quick ancillary test has the potential to prioritize cancer over benign tissues for expedited pathologic evaluation in poorly resourced countries.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. - PubMed
-
- Soerjomataram I, Lortet-Tieulent J, Parkin DM, Ferlay J, Mathers C, Forman D, et al. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet. 2012;380:1840–50. - PubMed
-
- Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–84. - PubMed
-
- Ezzati M, Pearson-Stuttard J, Bennett JE, Mathers CD. Acting on non-communicable diseases in low- and middle-income tropical countries. Nature. 2018;559:507–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

