CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes
- PMID: 31300537
- PMCID: PMC6681705
- DOI: 10.1073/pnas.1904714116
CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes
Abstract
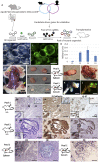
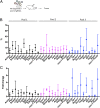
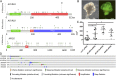
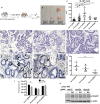
Colorectal cancer (CRC) is the third leading cause of cancer-related deaths worldwide. Several genome sequencing studies have provided comprehensive CRC genomic datasets. Likewise, in our previous study, we performed genome-wide Sleeping Beauty transposon-based mutagenesis screening in mice and provided comprehensive datasets of candidate CRC driver genes. However, functional validation for most candidate CRC driver genes, which were commonly identified from both human and mice, has not been performed. Here, we describe a platform for functionally validating CRC driver genes that utilizes CRISPR-Cas9 in mouse intestinal tumor organoids and human CRC-derived organoids in xenograft mouse models. We used genetically defined benign tumor-derived organoids carrying 2 frequent gene mutations (Apc and Kras mutations), which act in the early stage of CRC development, so that we could clearly evaluate the tumorigenic ability of the mutation in a single gene. These studies showed that Acvr1b, Acvr2a, and Arid2 could function as tumor suppressor genes (TSGs) in CRC and uncovered a role for Trp53 in tumor metastasis. We also showed that co-occurrent mutations in receptors for activin and transforming growth factor-β (TGF-β) synergistically promote tumorigenesis, and shed light on the role of activin receptors in CRC. This experimental system can also be applied to mouse intestinal organoids carrying other sensitizing mutations as well as organoids derived from other organs, which could further contribute to identification of novel cancer driver genes and new drug targets.
Keywords: CRISPR-Cas9; activin; colorectal cancer; driver gene; organoid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kinzler K. W., Vogelstein B., Lessons from hereditary colorectal cancer. Cell 87, 159–170 (1996). - PubMed
-
- Pinto D., Clevers H., Wnt, stem cells and cancer in the intestine. Biol. Cell 97, 185–196 (2005). - PubMed
-
- Bos J. L., ras oncogenes in human cancer: A review. Cancer Res. 49, 4682–4689 (1989). - PubMed
-
- Baker S. J., et al. , p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 50, 7717–7722 (1990). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

