PARP-1 and its associated nucleases in DNA damage response
- PMID: 31302005
- PMCID: PMC6764844
- DOI: 10.1016/j.dnarep.2019.102651
PARP-1 and its associated nucleases in DNA damage response
Abstract
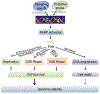
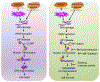
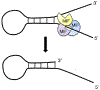
Poly(ADP-ribose) (PAR) polymerase-1 (PARP-1) acts as a DNA damage sensor. It recognizes DNA damage and facilitates DNA repair by recruiting DNA repair machinery to damage sites. Recent studies reported that PARP-1 also plays an important role in DNA replication by recognizing the unligated Okazaki fragments and controlling the speed of fork elongation. On the other hand, emerging evidence reveals that excessive activation of PARP-1 causes chromatin DNA fragmentation and triggers an intrinsic PARP-1-dependent cell death program designated parthanatos, which can be blocked by genetic deletion or pharmacological inhibition of PARP-1. Therefore, PARP-1 plays an essential role in maintaining genomic stability by either facilitating DNA repair/replication or triggering DNA fragmentation to kill cells. A group of structure-specific nucleases is crucial for executing DNA incision and fragmentation following PARP-1 activation. In this review, we will discuss how PARP-1 coordinates with its associated nucleases to maintain genomic integrity and control the decision of cell life and death.
Keywords: Cell death; DNA damage; DNA replication/repair; Nuclease; PARP-1.
Copyright © 2019 Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures

Similar articles
-
The Importance of Poly(ADP-Ribose) Polymerase as a Sensor of Unligated Okazaki Fragments during DNA Replication.Mol Cell. 2018 Jul 19;71(2):319-331.e3. doi: 10.1016/j.molcel.2018.06.004. Epub 2018 Jul 5. Mol Cell. 2018. PMID: 29983321 Free PMC article.
-
High speed of fork progression induces DNA replication stress and genomic instability.Nature. 2018 Jul;559(7713):279-284. doi: 10.1038/s41586-018-0261-5. Epub 2018 Jun 27. Nature. 2018. PMID: 29950726
-
Poly(ADP-ribose) Polymerase (PARP) and PARP Inhibitors: Mechanisms of Action and Role in Cardiovascular Disorders.Cardiovasc Toxicol. 2018 Dec;18(6):493-506. doi: 10.1007/s12012-018-9462-2. Cardiovasc Toxicol. 2018. PMID: 29968072 Review.
-
Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair.Methods Enzymol. 2006;409:493-510. doi: 10.1016/S0076-6879(05)09029-4. Methods Enzymol. 2006. PMID: 16793420
-
The comings and goings of PARP-1 in response to DNA damage.DNA Repair (Amst). 2018 Nov;71:177-182. doi: 10.1016/j.dnarep.2018.08.022. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30177435 Free PMC article. Review.
Cited by
-
Advances in the mechanism of small nucleolar RNA and its role in DNA damage response.Mil Med Res. 2024 Aug 8;11(1):53. doi: 10.1186/s40779-024-00553-4. Mil Med Res. 2024. PMID: 39118131 Free PMC article. Review.
-
Non-apoptotic regulated cell death in Fuchs endothelial corneal dystrophy.Regen Ther. 2023 Nov 10;24:592-601. doi: 10.1016/j.reth.2023.11.001. eCollection 2023 Dec. Regen Ther. 2023. PMID: 38034859 Free PMC article.
-
Role of PARP in TNBC: Mechanism of Inhibition, Clinical Applications, and Resistance.Biomedicines. 2021 Oct 21;9(11):1512. doi: 10.3390/biomedicines9111512. Biomedicines. 2021. PMID: 34829741 Free PMC article. Review.
-
PARP-1 and SIRT-1 are Interacted in Diabetic Nephropathy by Activating AMPK/PGC-1α Signaling Pathway.Diabetes Metab Syndr Obes. 2021 Jan 25;14:355-366. doi: 10.2147/DMSO.S291314. eCollection 2021. Diabetes Metab Syndr Obes. 2021. PMID: 33531822 Free PMC article.
-
Depletion of PARP10 inhibits the growth and metastatic potential of oral squamous cell carcinoma.Front Genet. 2022 Oct 13;13:1035638. doi: 10.3389/fgene.2022.1035638. eCollection 2022. Front Genet. 2022. PMID: 36313419 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

