A High-Throughput Screening Identifies MicroRNA Inhibitors That Influence Neuronal Maintenance and/or Response to Oxidative Stress
- PMID: 31302497
- PMCID: PMC6626867
- DOI: 10.1016/j.omtn.2019.06.007
A High-Throughput Screening Identifies MicroRNA Inhibitors That Influence Neuronal Maintenance and/or Response to Oxidative Stress
Abstract
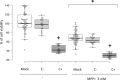
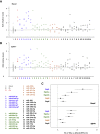
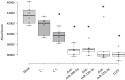
Small non-coding RNAs (sncRNAs), including microRNAs (miRNAs) are important post-transcriptional gene expression regulators relevant in physiological and pathological processes. Here, we combined a high-throughput functional screening (HTFS) platform with a library of antisense oligonucleotides (ASOs) to systematically identify sncRNAs that affect neuronal cell survival in basal conditions and in response to oxidative stress (OS), a major hallmark in neurodegenerative diseases. We considered hits commonly detected by two statistical methods in three biological replicates. Forty-seven ASOs targeting miRNAs (miRNA-ASOs) consistently decreased cell viability under basal conditions. A total of 60 miRNA-ASOs worsened cell viability impairment mediated by OS, with 36.6% commonly affecting cell viability under basal conditions. In addition, 40 miRNA-ASOs significantly protected neuronal cells from OS. In agreement with cell viability impairment, damaging miRNA-ASOs specifically induced increased free radical biogenesis. miRNAs targeted by the detrimental ASOs are enriched in the fraction of miRNAs downregulated by OS, suggesting that the miRNA expression pattern after OS contributes to neuronal damage. The present HTFS highlighted potentially druggable sncRNAs. However, future studies are needed to define the pathways by which the identified ASOs regulate cell survival and OS response and to explore the potential of translating the current findings into clinical applications.
Keywords: expression profiles; high-throughput screening; miRNAs; mitochondrial function; neurodegeneration; non-coding RNAs; oxidative stress; small RNA sequencing.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Post-transcriptional gene-expression regulation by micro RNA (miRNA) network in renal disease.Adv Drug Deliv Rev. 2010 Nov 30;62(14):1390-401. doi: 10.1016/j.addr.2010.10.003. Epub 2010 Oct 19. Adv Drug Deliv Rev. 2010. PMID: 20940025 Review.
-
Use of steric blocking antisense oligonucleotides for the targeted inhibition of junction containing precursor microRNAs.bioRxiv [Preprint]. 2024 Apr 8:2024.04.08.588531. doi: 10.1101/2024.04.08.588531. bioRxiv. 2024. PMID: 38645194 Free PMC article. Preprint.
-
Oxidative stress modulates the expression of apoptosis-associated microRNAs in bovine granulosa cells in vitro.Cell Tissue Res. 2019 May;376(2):295-308. doi: 10.1007/s00441-019-02990-3. Epub 2019 Jan 21. Cell Tissue Res. 2019. PMID: 30666538
-
The Emerging Role of MitomiRs in the Pathophysiology of Human Disease.Adv Exp Med Biol. 2015;888:123-54. doi: 10.1007/978-3-319-22671-2_8. Adv Exp Med Biol. 2015. PMID: 26663182
-
Role of microRNA (miRNA) and Viroids in Lethal Diseases of Plants and Animals. Potential Contribution to Human Neurodegenerative Disorders.Biochemistry (Mosc). 2018 Sep;83(9):1018-1029. doi: 10.1134/S0006297918090031. Biochemistry (Mosc). 2018. PMID: 30472940 Review.
Cited by
-
Disease- and headache-specific microRNA signatures and their predicted mRNA targets in peripheral blood mononuclear cells in migraineurs: role of inflammatory signalling and oxidative stress.J Headache Pain. 2022 Sep 2;23(1):113. doi: 10.1186/s10194-022-01478-w. J Headache Pain. 2022. PMID: 36050647 Free PMC article.
-
Construction and Investigation of MicroRNA-mRNA Regulatory Network of Gastric Cancer with Helicobacter pylori Infection.Biochem Res Int. 2020 Jul 25;2020:6285987. doi: 10.1155/2020/6285987. eCollection 2020. Biochem Res Int. 2020. PMID: 32802507 Free PMC article.
-
Exploring Common Therapeutic Targets for Neurodegenerative Disorders Using Transcriptome Study.Front Genet. 2021 Mar 19;12:639160. doi: 10.3389/fgene.2021.639160. eCollection 2021. Front Genet. 2021. PMID: 33815473 Free PMC article.
-
Designing libraries for pooled CRISPR functional screens of long noncoding RNAs.Mamm Genome. 2022 Jun;33(2):312-327. doi: 10.1007/s00335-021-09918-9. Epub 2021 Sep 17. Mamm Genome. 2022. PMID: 34533605 Free PMC article. Review.
-
Identifying a novel biological mechanism for alcohol addiction associated with circRNA networks acting as potential miRNA sponges.Addict Biol. 2021 Nov;26(6):e13071. doi: 10.1111/adb.13071. Epub 2021 Jun 23. Addict Biol. 2021. PMID: 34164896 Free PMC article.
References
-
- Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019;20:5–20. - PubMed
-
- Cao X., Yeo G., Muotri A.R., Kuwabara T., Gage F.H. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006;29:77–103. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

