Mutations in RHOT1 Disrupt Endoplasmic Reticulum-Mitochondria Contact Sites Interfering with Calcium Homeostasis and Mitochondrial Dynamics in Parkinson's Disease
- PMID: 31303019
- PMCID: PMC6798875
- DOI: 10.1089/ars.2018.7718
Mutations in RHOT1 Disrupt Endoplasmic Reticulum-Mitochondria Contact Sites Interfering with Calcium Homeostasis and Mitochondrial Dynamics in Parkinson's Disease
Erratum in
-
Correction to: Mutations in RHOT1 Disrupt Endoplasmic Reticulum-Mitochondria Contact Sites Interfering with Calcium Homeostasis and Mitochondrial Dynamics in Parkinson's Disease by Grossmann D, Berenguer-Escuder C, Bellet ME, Scheibner D, Bohler J, Massart F, Rapaport D, Skupin A, Fouquier d'Hérouël A, Sharma M, Ghelfi J, Raković A, Lichtner P, Antony P, Glaab E, May P, Dimmer KS, Fitzgerald JC, Grünewald A, and Krüger R Antioxid Redox Signal 31: 1213-1234, 2019. DOI: 10.1089/ars.2018.7718.Antioxid Redox Signal. 2020 Aug 12;33(11):786-7. doi: 10.1089/ars.2018.7718.correx. Online ahead of print. Antioxid Redox Signal. 2020. PMID: 32835496 Free PMC article. No abstract available.
Abstract
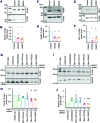
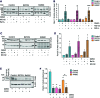
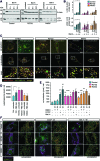
Aims: The outer mitochondrial membrane protein Miro1 is a crucial player in mitochondrial dynamics and calcium homeostasis. Recent evidence indicated that Miro1 mediates calcium-induced mitochondrial shape transition, which is a prerequisite for the initiation of mitophagy. Moreover, altered Miro1 protein levels have emerged as a shared feature of monogenic and sporadic Parkinson's disease (PD), but, so far, no disease-associated variants in RHOT1 have been identified. Here, we aim to explore the genetic and functional contribution of RHOT1 mutations to PD in patient-derived cellular models. Results: For the first time, we describe heterozygous RHOT1 mutations in two PD patients (het c.815G>A; het c.1348C>T) and identified mitochondrial phenotypes with reduced mitochondrial mass in patient fibroblasts. Both mutations led to decreased endoplasmic reticulum-mitochondrial contact sites and calcium dyshomeostasis. As a consequence, energy metabolism was impaired, which in turn caused increased mitophagy. Innovation and Conclusion: Our study provides functional evidence that ROTH1 is a genetic risk factor for PD, further implicating Miro1 in calcium homeostasis and mitochondrial quality control.
Keywords: ER–mitochondria contact site; Miro1; Parkinson's disease; calcium; mitochondria; patient fibroblasts.
Conflict of interest statement
R.K. received honoraria/travel grants from Abbvie, Zambon, and Medtronic and participated as site Principal Investigator for industry-sponsored clinical trials without receiving additional honoraria. All other authors have no competing financial interests.
Figures




Similar articles
-
The Emerging Role of RHOT1/Miro1 in the Pathogenesis of Parkinson's Disease.Front Neurol. 2020 Sep 15;11:587. doi: 10.3389/fneur.2020.00587. eCollection 2020. Front Neurol. 2020. PMID: 33041957 Free PMC article. Review.
-
Impaired mitochondrial-endoplasmic reticulum interaction and mitophagy in Miro1-mutant neurons in Parkinson's disease.Hum Mol Genet. 2020 May 28;29(8):1353-1364. doi: 10.1093/hmg/ddaa066. Hum Mol Genet. 2020. PMID: 32280985 Free PMC article.
-
Variants in Miro1 Cause Alterations of ER-Mitochondria Contact Sites in Fibroblasts from Parkinson's Disease Patients.J Clin Med. 2019 Dec 16;8(12):2226. doi: 10.3390/jcm8122226. J Clin Med. 2019. PMID: 31888276 Free PMC article.
-
Generation of two induced pluripotent stem cell lines and the corresponding isogenic controls from Parkinson's disease patients carrying the heterozygous mutations c.815G > A (p.R272Q) or c.1348C > T (p.R450C) in the RHOT1 gene encoding Miro1.Stem Cell Res. 2023 Sep;71:103145. doi: 10.1016/j.scr.2023.103145. Epub 2023 Jun 14. Stem Cell Res. 2023. PMID: 37364399
-
Mitochondrial-Associated Membranes in Parkinson's Disease.Adv Exp Med Biol. 2017;997:157-169. doi: 10.1007/978-981-10-4567-7_12. Adv Exp Med Biol. 2017. PMID: 28815529 Review.
Cited by
-
The Molecular Determinants of Mitochondrial Membrane Contact With ER, Lysosomes and Peroxisomes in Neuronal Physiology and Pathology.Front Cell Neurosci. 2020 Aug 7;14:194. doi: 10.3389/fncel.2020.00194. eCollection 2020. Front Cell Neurosci. 2020. PMID: 32848610 Free PMC article.
-
Loss of neuronal Miro1 disrupts mitophagy and induces hyperactivation of the integrated stress response.EMBO J. 2021 Jul 15;40(14):e100715. doi: 10.15252/embj.2018100715. Epub 2021 Jun 21. EMBO J. 2021. PMID: 34152608 Free PMC article.
-
The Emerging Role of RHOT1/Miro1 in the Pathogenesis of Parkinson's Disease.Front Neurol. 2020 Sep 15;11:587. doi: 10.3389/fneur.2020.00587. eCollection 2020. Front Neurol. 2020. PMID: 33041957 Free PMC article. Review.
-
The Mitochondrial-Associated Endoplasmic Reticulum Membrane and Its Role in Diabetic Nephropathy.Oxid Med Cell Longev. 2021 Nov 5;2021:8054817. doi: 10.1155/2021/8054817. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34777695 Free PMC article. Review.
-
Calcium Homeostasis in Parkinson's Disease: From Pathology to Treatment.Neurosci Bull. 2022 Oct;38(10):1267-1270. doi: 10.1007/s12264-022-00899-6. Epub 2022 Jun 21. Neurosci Bull. 2022. PMID: 35727497 Free PMC article. No abstract available.
References
-
- Basso V, Marchesan E, Peggion C, Chakraborty J, von Stockum S, Giacomello M, Ottolini D, Debattisti V, Caicci F, Tasca E, Pegoraro V, Angelini C, Antonini A, Bertoli A, Brini M, and Ziviani E. Regulation of ER-mitochondria contacts by Parkin via Mfn2. Pharmacol Res 138: 43–56, 2018 - PubMed
-
- Birsa N, Norkett R, Wauer T, Mevissen TET, Wu HC, Foltynie T, Bhatia K, Hirst WD, Komander D, Plun-Favreau H, and Kittler JT. Lysine 27 ubiquitination of the mitochondrial transport protein miro is dependent on serine 65 of the parkin ubiquitin ligase. J Biol Chem 289: 14569–14582, 2014 - PMC - PubMed
-
- Burbulla LF, Schelling C, Kato H, Rapaport D, Woitalla D, Schiesling C, Schulte C, Sharma M, Illig T, Bauer P, Jung S, Nordheim A, Schöls L, Riess O, and Krüger R. Dissecting the role of the mitochondrial chaperone mortalin in Parkinson's disease: functional impact of disease-related variants on mitochondrial homeostasis. Hum Mol Genet 19: 4437–4452, 2010 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
