Replisome structure suggests mechanism for continuous fork progression and post-replication repair
- PMID: 31303546
- PMCID: PMC7467748
- DOI: 10.1016/j.dnarep.2019.102658
Replisome structure suggests mechanism for continuous fork progression and post-replication repair
Abstract
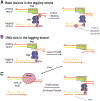
What happens to DNA replication when it encounters a damaged or nicked DNA template has been under investigation for five decades. Initially it was thought that DNA polymerase, and thus the replication-fork progression, would stall at road blocks. After the discovery of replication-fork helicase and replication re-initiation factors by the 1990s, it became clear that the replisome can "skip" impasses and finish replication with single-stranded gaps and double-strand breaks in the product DNA. But the mechanism for continuous fork progression after encountering roadblocks is entangled with translesion synthesis, replication fork reversal and recombination repair. The recently determined structure of the bacteriophage T7 replisome offers the first glimpse of how helicase, primase, leading-and lagging-strand DNA polymerases are organized around a DNA replication fork. The tightly coupled leading-strand polymerase and lagging-strand helicase provides a scaffold to consolidate data accumulated over the past five decades and offers a fresh perspective on how the replisome may skip lesions and complete discontinuous DNA synthesis. Comparison of the independently evolved bacterial and eukaryotic replisomes suggests that repair of discontinuous DNA synthesis occurs post replication in both.
Keywords: Helicase reload; Lesion skipping; Polymerase restart; Replication fork; Replisome.
Copyright © 2019. Published by Elsevier B.V.
Figures

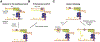

References
-
- Watson JD, Crick FH, The structure of DNA, Cold Spring Harb. Symp. Quant. Biol 18 (1953) 123–131. - PubMed
-
- Bessman MJ, Kornberg A, Lehman IR, Simms ES, Enzymic synthesis of deoxyribonucleic acid, Biochim. Biophys. Acta 21 (1956) 197–198. - PubMed
-
- Merz T, Swanson CP, Hemalatha CN, Radiosensitivity and problem of chromosome breakage and rejoining, Brookhaven Symp. Biol 14 (1961) 53–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

