Generation of Human PSC-Derived Kidney Organoids with Patterned Nephron Segments and a De Novo Vascular Network
- PMID: 31303547
- PMCID: PMC6731150
- DOI: 10.1016/j.stem.2019.06.009
Generation of Human PSC-Derived Kidney Organoids with Patterned Nephron Segments and a De Novo Vascular Network
Abstract
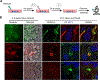
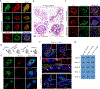
Human pluripotent stem cell-derived kidney organoids recapitulate developmental processes and tissue architecture, but intrinsic limitations, such as lack of vasculature and functionality, have greatly hampered their application. Here we establish a versatile protocol for generating vascularized three-dimensional (3D) kidney organoids. We employ dynamic modulation of WNT signaling to control the relative proportion of proximal versus distal nephron segments, producing a correlative level of vascular endothelial growth factor A (VEGFA) to define a resident vascular network. Single-cell RNA sequencing identifies a subset of nephron progenitor cells as a potential source of renal vasculature. These kidney organoids undergo further structural and functional maturation upon implantation. Using this kidney organoid platform, we establish an in vitro model of autosomal recessive polycystic kidney disease (ARPKD), the cystic phenotype of which can be effectively prevented by gene correction or drug treatment. Our studies provide new avenues for studying human kidney development, modeling disease pathogenesis, and performing patient-specific drug validation.
Keywords: ARPKD; developmental modeling; disease modeling; implantation; kidney organoid; nephron patterning; patient-specific iPSC; pluripotent stem cells; single-cell RNA-seq; vascular progenitors.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





References
-
- ABRAHAMSON DR, ROBERT B, HYINK DP, ST JOHN PL & DANIEL TO 1998. Origins and formation of microvasculature in the developing kidney. Kidney Int Suppl, 67, S7–11. - PubMed
-
- BARASCH J, YANG J, WARE CB, TAGA T, YOSHIDA K, ERDJUMENT-BROMAGE H, TEMPST P, PARRAVICINI E, MALACH S, ARANOFF T & OLIVER JA 1999. Mesenchymal to epithelial conversion in rat metanephros is induced by LIF. Cell, 99, 377–86. - PubMed
-
- BIRN H & CHRISTENSEN EI 2006. Renal albumin absorption in physiology and pathology. Kidney Int, 69, 440–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

