Endogenous and Foreign Nucleoid-Associated Proteins of Bacteria: Occurrence, Interactions and Effects on Mobile Genetic Elements and Host's Biology
- PMID: 31303979
- PMCID: PMC6606824
- DOI: 10.1016/j.csbj.2019.06.010
Endogenous and Foreign Nucleoid-Associated Proteins of Bacteria: Occurrence, Interactions and Effects on Mobile Genetic Elements and Host's Biology
Abstract
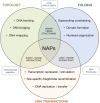
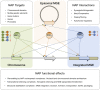
Mobile Genetic Elements (MGEs) are mosaics of functional gene modules of diverse evolutionary origin and are generally divergent from the hosts´ genetic background. Existing biases in base composition and codon usage of these elements` genes impose transcription and translation limitations that may affect the physical and regulatory integration of MGEs in new hosts. Stable appropriation of the foreign DNA depends on a number of host factors among which are the Nucleoid-Associated Proteins (NAPs). These small, basic, highly abundant proteins bind and bend DNA, altering its topology and folding, thereby affecting all known essential DNA metabolism related processes. Both chromosomally- (endogenous) and MGE- (foreign) encoded NAPs have been shown to exist in bacteria. While the role of host-encoded NAPs in xenogeneic silencing of both episomal (plasmids) and integrative MGEs (pathogenicity islands and prophages) is well acknowledged, less is known about the role of MGE-encoded NAPs in the foreign elements biology or their influence on the host's chromosome expression dynamics. Here we review existing literature on the topic, present examples on the positive and negative effects that endogenous and foreign NAPs exert on global transcriptional gene expression, MGE integrative and excisive recombination dynamics, persistence and transfer to suitable hosts and discuss the nature and relevance of synergistic and antagonizing higher order interactions between diverse types of NAPs.
Keywords: Bacterial nucleoid; HGT; MGE; NAP; Regulatory hierarchies; Xenogeneic silencing.
Figures
Similar articles
-
Pangenome-level analysis of nucleoid-associated proteins in the Acidithiobacillia class: insights into their functional roles in mobile genetic elements biology.Front Microbiol. 2023 Sep 25;14:1271138. doi: 10.3389/fmicb.2023.1271138. eCollection 2023. Front Microbiol. 2023. PMID: 37817747 Free PMC article.
-
mobileOG-db: a Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements.Appl Environ Microbiol. 2022 Sep 22;88(18):e0099122. doi: 10.1128/aem.00991-22. Epub 2022 Aug 29. Appl Environ Microbiol. 2022. PMID: 36036594 Free PMC article.
-
Bacterial nucleoid-associated proteins, nucleoid structure and gene expression.Nat Rev Microbiol. 2010 Mar;8(3):185-95. doi: 10.1038/nrmicro2261. Epub 2010 Feb 8. Nat Rev Microbiol. 2010. PMID: 20140026 Review.
-
Nucleoid-associated proteins encoded on plasmids: Occurrence and mode of function.Plasmid. 2015 Jul;80:32-44. doi: 10.1016/j.plasmid.2015.04.008. Epub 2015 May 4. Plasmid. 2015. PMID: 25952329
-
H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria.Plasmid. 2014 Sep;75:1-11. doi: 10.1016/j.plasmid.2014.06.004. Epub 2014 Jul 3. Plasmid. 2014. PMID: 24998344 Review.
Cited by
-
Machine Learning Suggests That Small Size Helps Broaden Plasmid Host Range.Genes (Basel). 2023 Nov 5;14(11):2044. doi: 10.3390/genes14112044. Genes (Basel). 2023. PMID: 38002987 Free PMC article.
-
Genome and sequence determinants governing the expression of horizontally acquired DNA in bacteria.ISME J. 2020 Sep;14(9):2347-2357. doi: 10.1038/s41396-020-0696-1. Epub 2020 Jun 8. ISME J. 2020. PMID: 32514119 Free PMC article.
-
Nucleoid-associated proteins: molecular mechanisms in microbial adaptation.World J Microbiol Biotechnol. 2025 Jul 28;41(8):277. doi: 10.1007/s11274-025-04419-2. World J Microbiol Biotechnol. 2025. PMID: 40719955 Review.
-
Novel anti-repression mechanism of H-NS proteins by a phage protein.Nucleic Acids Res. 2021 Oct 11;49(18):10770-10784. doi: 10.1093/nar/gkab793. Nucleic Acids Res. 2021. PMID: 34520554 Free PMC article.
-
Nucleoid Associated Proteins: The Small Organizers That Help to Cope With Stress.Front Microbiol. 2020 Apr 8;11:590. doi: 10.3389/fmicb.2020.00590. eCollection 2020. Front Microbiol. 2020. PMID: 32373086 Free PMC article. Review.
References
-
- Ali Azam T.A., Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. - PubMed
-
- Auner H., Buckle M., Deufel A., Kutateladze T., Lazarus L. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J Mol Biol. 2003;331:331–344. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
