Within-host infectious disease models accommodating cellular coinfection, with an application to influenza
- PMID: 31304043
- PMCID: PMC6613536
- DOI: 10.1093/ve/vez018
Within-host infectious disease models accommodating cellular coinfection, with an application to influenza
Erratum in
-
Erratum: Santa Fe Institute Workshop Special Issue articles.Virus Evol. 2019 Nov 19;5(2):vez052. doi: 10.1093/ve/vez052. eCollection 2019 Jul. Virus Evol. 2019. PMID: 31768266 Free PMC article.
Abstract
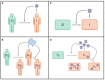
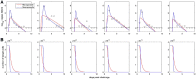
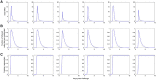
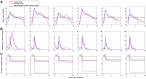
Within-host models are useful tools for understanding the processes regulating viral load dynamics. While existing models have considered a wide range of within-host processes, at their core these models have shown remarkable structural similarity. Specifically, the structure of these models generally consider target cells to be either uninfected or infected, with the possibility of accommodating further resolution (e.g. cells that are in an eclipse phase). Recent findings, however, indicate that cellular coinfection is the norm rather than the exception for many viral infectious diseases, and that cells with high multiplicity of infection are present over at least some duration of an infection. The reality of these cellular coinfection dynamics is not accommodated in current within-host models although it may be critical for understanding within-host dynamics. This is particularly the case if multiplicity of infection impacts infected cell phenotypes such as their death rate and their viral production rates. Here, we present a new class of within-host disease models that allow for cellular coinfection in a scalable manner by retaining the low-dimensionality that is a desirable feature of many current within-host models. The models we propose adopt the general structure of epidemiological 'macroparasite' models that allow hosts to be variably infected by parasites such as nematodes and host phenotypes to flexibly depend on parasite burden. Specifically, our within-host models consider target cells as 'hosts' and viral particles as 'macroparasites', and allow viral output and infected cell lifespans, among other phenotypes, to depend on a cell's multiplicity of infection. We show with an application to influenza that these models can be statistically fit to viral load and other within-host data, and demonstrate using model selection approaches that they have the ability to outperform traditional within-host viral dynamic models. Important in vivo quantities such as the mean multiplicity of cellular infection and time-evolving reassortant frequencies can also be quantified in a straightforward manner once these macroparasite models have been parameterized. The within-host model structure we develop here provides a mathematical way forward to address questions related to the roles of cellular coinfection, collective viral interactions, and viral complementation in within-host viral dynamics and evolution.
Keywords: cellular coinfection; influenza virus; macroparasite model; viral complementation; within-host dynamics.
Figures
References
-
- Anderson R. M., May R. M. (1978) ‘Regulation and Stability of Host-Parasite Population Interactions: I. Regulatory Processes’, The Journal of Animal Ecology, 47: 249–67.
-
- Anderson R. M., May R. M. (1992) Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford University Press.
Grants and funding
LinkOut - more resources
Full Text Sources

