Comprehensive analysis of an lncRNA-miRNA-mRNA competing endogenous RNA network in pulpitis
- PMID: 31304055
- PMCID: PMC6609876
- DOI: 10.7717/peerj.7135
Comprehensive analysis of an lncRNA-miRNA-mRNA competing endogenous RNA network in pulpitis
Abstract
Background: Pulpitis is a common inflammatory disease that affects dental pulp. It is important to understand the molecular signals of inflammation and repair associated with this process. Increasing evidence has revealed that long noncoding RNAs (lncRNAs), via competitively sponging microRNAs (miRNAs), can act as competing endogenous RNAs (ceRNAs) to regulate inflammation and reparative responses. The aim of this study was to elucidate the potential roles of lncRNA, miRNA and messenger RNA (mRNA) ceRNA networks in pulpitis tissues compared to normal control tissues.
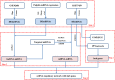
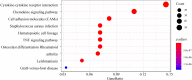
Methods: The oligo and limma packages were used to identify differentially expressed lncRNAs and mRNAs (DElncRNAs and DEmRNAs, respectively) based on expression profiles in two datasets, GSE92681 and GSE77459, from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) were further analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Protein-protein interaction (PPI) networks and modules were established to screen hub genes using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) and the Molecular Complex Detection (MCODE) plugin for Cytoscape, respectively. Furthermore, an lncRNA-miRNA-mRNA-hub genes regulatory network was constructed to investigate mechanisms related to the progression and prognosis of pulpitis. Then, quantitative real-time polymerase chain reaction (qRT-PCR) was applied to verify critical lncRNAs that may significantly affect the pathogenesis in inflamed and normal human dental pulp.
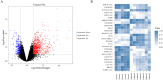
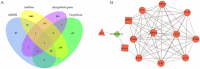
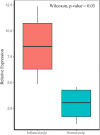
Results: A total of 644 upregulated and 264 downregulated differentially expressed genes (DEGs) in pulpitis samples were identified from the GSE77459 dataset, while 8 up- and 19 downregulated probes associated with lncRNA were identified from the GSE92681 dataset. Protein-protein interaction (PPI) based on STRING analysis revealed a network of DEGs containing 4,929 edges and 623 nodes. Upon combined analysis of the constructed PPI network and the MCODE results, 10 hub genes, including IL6, IL8, PTPRC, IL1B, TLR2, ITGAM, CCL2, PIK3CG, ICAM1, and PIK3CD, were detected in the network. Next, a ceRNA regulatory relationship consisting of one lncRNA (PVT1), one miRNA (hsa-miR-455-5p) and two mRNAs (SOCS3 and PLXNC1) was established. Then, we constructed the network in which the regulatory relationship between ceRNA and hub genes was summarized. Finally, our qRT-PCR results confirmed significantly higher levels of PVT1 transcript in inflamed pulp than in normal pulp tissues (p = 0.03).
Conclusion: Our study identified a novel lncRNA-mediated ceRNA regulatory mechanisms in the pathogenesis of pulpitis.
Keywords: Bioinformatics analysis; Competing endogenous RNA network; Long noncoding RNA; Pulpitis.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Bioinformatics Analysis of the Mechanisms of Diabetic Nephropathy via Novel Biomarkers and Competing Endogenous RNA Network.Front Endocrinol (Lausanne). 2022 Jul 14;13:934022. doi: 10.3389/fendo.2022.934022. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35909518 Free PMC article.
-
Exploring the Molecular Mechanism of lncRNA-miRNA-mRNA Networks in Non-Syndromic Cleft Lip with or without Cleft Palate.Int J Gen Med. 2021 Dec 16;14:9931-9943. doi: 10.2147/IJGM.S339504. eCollection 2021. Int J Gen Med. 2021. PMID: 34938111 Free PMC article.
-
Identification of a lncRNA/circRNA-miRNA-mRNA ceRNA Network in Alzheimer's Disease.J Integr Neurosci. 2023 Oct 17;22(6):136. doi: 10.31083/j.jin2206136. J Integr Neurosci. 2023. PMID: 38176923
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
Bioinformatics analysis of ceRNA network of autophagy-related genes in pediatric asthma.Medicine (Baltimore). 2023 Dec 1;102(48):e36343. doi: 10.1097/MD.0000000000036343. Medicine (Baltimore). 2023. PMID: 38050261 Free PMC article. Review.
Cited by
-
A novel NUTM2A-AS1/miR-769-5p axis regulates LPS-evoked damage in human dental pulp cells via the TLR4/MYD88/NF-κB signaling.J Dent Sci. 2025 Jul;20(3):1579-1589. doi: 10.1016/j.jds.2022.05.010. Epub 2022 Jun 4. J Dent Sci. 2025. PMID: 40654486 Free PMC article.
-
Wnt4 prevents apoptosis and inflammation of dental pulp cells induced by LPS by inhibiting the IKK/NF‑κB pathway.Exp Ther Med. 2022 Dec 23;25(2):75. doi: 10.3892/etm.2022.11774. eCollection 2023 Feb. Exp Ther Med. 2022. PMID: 36684653 Free PMC article.
-
Diagnostic biomarker candidates for pulpitis revealed by bioinformatics analysis of merged microarray gene expression datasets.BMC Oral Health. 2020 Oct 12;20(1):279. doi: 10.1186/s12903-020-01266-5. BMC Oral Health. 2020. PMID: 33046027 Free PMC article.
-
Dysregulation of MicroRNA-152-3p is Associated with the Pathogenesis of Pulpitis by Modulating SMAD5.Oral Health Prev Dent. 2023 Jun 5;21:211-218. doi: 10.3290/j.ohpd.b4132867. Oral Health Prev Dent. 2023. PMID: 37272598 Free PMC article.
-
Long Noncoding RNA MIR4435-2HG is Involved in Osteogenic Differentiation and Inflammation in Human Dental Pulp Stem Cells.Int Dent J. 2025 Aug;75(4):100866. doi: 10.1016/j.identj.2025.100866. Epub 2025 Jun 25. Int Dent J. 2025. PMID: 40570674 Free PMC article.
References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

