Transcriptional regulation of normal human mammary cell heterogeneity and its perturbation in breast cancer
- PMID: 31304632
- PMCID: PMC6627240
- DOI: 10.15252/embj.2018100330
Transcriptional regulation of normal human mammary cell heterogeneity and its perturbation in breast cancer
Abstract
The mammary gland in adult women consists of biologically distinct cell types that differ in their surface phenotypes. Isolation and molecular characterization of these subpopulations of mammary cells have provided extensive insights into their different transcriptional programs and regulation. This information is now serving as a baseline for interpreting the heterogeneous features of human breast cancers. Examination of breast cancer mutational profiles further indicates that most have undergone a complex evolutionary process even before being detected. The consequent intra-tumoral as well as inter-tumoral heterogeneity of these cancers thus poses major challenges to deriving information from early and hence likely pervasive changes in potential therapeutic interest. Recently described reproducible and efficient methods for generating human breast cancers de novo in immunodeficient mice transplanted with genetically altered primary cells now offer a promising alternative to investigate initial stages of human breast cancer development. In this review, we summarize current knowledge about key transcriptional regulatory processes operative in these partially characterized subpopulations of normal human mammary cells and effects of disrupting these processes in experimentally produced human breast cancers.
Keywords: breast cancer; chromatin; epigenomics; mammary; transcriptional regulation.
© 2019 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


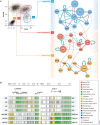
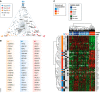
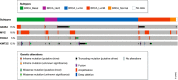
References
-
- Aboussekhra A (2011) Role of cancer‐associated fibroblasts in breast cancer development and prognosis. Int J Dev Biol 55: 841–849 - PubMed
-
- Adams JR, Xu K, Liu JC, Agamez NMR, Loch AJ, Wong RG, Wang W, Wright KL, Lane TF, Zacksenhaus E, Egan SE (2011) Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res 71: 2706–2717 - PubMed
-
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti‐Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K (2004) Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 6: 17–32 - PubMed
-
- van Amerongen R, Bowman AN, Nusse R (2012) Developmental stage and time dictate the fate of Wnt/β‐catenin‐responsive stem cells in the mammary gland. Cell Stem Cell 11: 387–400 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

