Conserved crosstalk between histone deacetylation and H3K79 methylation generates DOT1L-dose dependency in HDAC1-deficient thymic lymphoma
- PMID: 31304633
- PMCID: PMC6627229
- DOI: 10.15252/embj.2019101564
Conserved crosstalk between histone deacetylation and H3K79 methylation generates DOT1L-dose dependency in HDAC1-deficient thymic lymphoma
Abstract
DOT1L methylates histone H3K79 and is aberrantly regulated in MLL-rearranged leukemia. Inhibitors have been developed to target DOT1L activity in leukemia, but cellular mechanisms that regulate DOT1L are still poorly understood. We have identified the histone deacetylase Rpd3 as a negative regulator of budding yeast Dot1. At its target genes, the transcriptional repressor Rpd3 restricts H3K79 methylation, explaining the absence of H3K79me3 at a subset of genes in the yeast genome. Similar to the crosstalk in yeast, inactivation of the murine Rpd3 homolog HDAC1 in thymocytes led to an increase in H3K79 methylation. Thymic lymphomas that arise upon genetic deletion of Hdac1 retained the increased H3K79 methylation and were sensitive to reduced DOT1L dosage. Furthermore, cell lines derived from Hdac1Δ/Δ thymic lymphomas were sensitive to a DOT1L inhibitor, which induced apoptosis. In summary, we identified an evolutionarily conserved crosstalk between HDAC1 and DOT1L with impact in murine thymic lymphoma development.
Keywords: Chromatin; H3K79 methylation; histone acetylation; histone ubiquitination; lymphoma.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The Netherlands Cancer Institute and FvL are entitled to royalties that may result from licensing the yeast H2BK123ub‐specific monoclonal antibody according to IP policies of the Netherlands Cancer Institute. The other authors declare that they have no conflict of interest.
Figures

Schematic overview of the Epi‐ID strategy.
Epi‐ID H3K79 methylation scores of the deletion mutants of members of the Rpd3L and Rpd3S complexes, calculated as described in Appendix Supplementary Methods, where 0 means a wild‐type H3K79me level (log2 scale). The gray dots indicate accessory subunits. UpTag and DownTag are barcode reporters in a promoter and terminator context, respectively. Data were obtained on all Rpd3L/Rpd3S subunits except Sds3.
Mass spectrometry analysis of H3K79 methylation in wild‐type and mutant strains. Mean and individual data points of two biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001 by two‐way ANOVA, comparison to wild type.
Heatmaps of H3K79me1, H3K79me3, H2Bub, and H3 in wild‐type cells, aligned on the TSS. Genes were sorted based on the average H3K79me3/H3K79me1 ratio in the first 500 bp.
Snapshot of depth‐normalized ChIP‐seq data tracks from wild‐type and rpd3Δ strains showing 6 kb surrounding the DBP1 ORF, which is the top gene in the heatmap in panel (F). All tracks have the same y‐axis (0–20 rpm). A snapshot of another top‐regulated gene is shown in Fig EV1D.
Heatmap of the H3K79me3/H3 change in rpd3Δ versus wild‐type cells, aligned on the TSS. Genes were sorted based on the average ratio in the first 500 bp.

- A
Immunoblots show that deletion of RPD3 or SIN3 does not lead to a detectable increase in global H2Bub or Dot1 protein levels.
- B
Metagene plots of H3K79me1, H3K79me3, H2Bub, H3, and H2B in rpd3Δ and wild‐type strains.
- C
Gene set enrichment analysis shows that subtelomeric genes (< 30 kb of telomeres) are enriched among genes with low H3K79 methylation (measured by the average H3K79me3/H3K79me1 ratio in the first 500 bp of the ORF).
- D–E
Snapshots of depth‐normalized ChIP‐seq data tracks from wild‐type and rpd3Δ strains showing 7 kb surrounding meiotic gene ZIP1 (D) and subtelomeric genes AIF1 and COS10 (E). All tracks have the same y‐axis (0–20 rpm), which, for comparison, is also the same scale as in Fig 1E.

- A
ChIP‐seq and RNA‐seq data for genes ranked on H3K79me3/H3 in rpd3Δ/WT, smoothed using locally weighted regression. The gray band around the line shows the 95% confidence interval. Vertical dashed lines separate 4 groups with distinct changes upon RPD3 deletion. ChIP‐seq data of H3K79me1, H3K79me3, and H2Bub were generated in this study (plotted is the average coverage in reads per genomic content, RPGC), Rpd3 binding, H4ac, and WT gene expression data were from McKnight et al (2015), and the relative expression in rpd3Δ/WT was from Kemmeren et al (2014).
- B
H3K79me3/H3 ChIP‐qPCR efficiencies (relative to a non‐transcribed region, which was unaffected by RPD3 deletion) in wild‐type and in rpd3Δ cells harboring empty or RPD3‐encoding CEN plasmids. The H188A and H150A‐H151A mutations have previously been shown to abrogate catalytic activity (Kadosh & Struhl, 1998). Error bars indicate standard deviation of three biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001 by two‐way ANOVA, comparison to wild type.
- C–E
Gene set enrichment analysis on genes ranked on H3K79me3/H3 in rpd3Δ/WT; all genes have been ranked, and the ranks of the genes in the subsets are indicated by vertical lines. Meiotic (C) and Ume6‐bound (D) genes are enriched among the genes at which Rpd3 represses H3K79 methylation, and subtelomeric genes (<30 kb of telomere) (E) are enriched among genes at which H3K79 methylation is decreased in rpd3Δ cells.

Heatmaps of H3K79me1, H3K79me3, H2Bub, and H3 sorted on H3K79me3/H3 rpd3Δ/WT.
H2Bub ChIP‐qPCR (H2Bub/H2B) at 5′ ends of indicated genes in wild‐type and rpd3Δ cells, with bre1Δ cells serving as a negative control. Error bars indicate standard deviation of three biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001 by two‐way ANOVA, comparison to wild type.
H2Bub ChIP‐qPCR (H2Bub/input) at the subtelomeric IRC7 gene, the promoter of the barcoded HO locus, and a non‐transcribed region (NoORF) (Imbeault et al, 2008; Verzijlbergen et al, 2010). Error bars indicate standard deviation of three biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001 by two‐way ANOVA, comparison to wild type.
ChIP‐seq and RNA‐seq data per gene (same data as in Fig 2A) ranked on gene expression level in wild‐type cells, smoothed using locally weighted regression. The shaded band around the line shows the 95% confidence interval.
Similar to panel (D), but using a ranking based on the level of antisense transcription per gene in wild‐type cells as calculated by Brown et al (2018) using data from Churchman and Weissman (2011).
H3K79me3 ChIP‐seq data in gene promoters (−400 to TSS, where the Arp5 effect is maximal) in wild‐type and arp5Δ cells (Xue et al, 2015), ranked by the effect Rpd3 has on H3K79me3 on each gene (same ranking as in Fig 2A).

Mass spectrometry analysis of H3K79 methylation in thymuses from 3‐week‐old mice, either wild‐type (Cre‐) or with deleted Hdac1 or Dot1L alleles, as indicated. Mean and individual data points of biological replicates; H3K79me0 is the predominant state, and the y‐axis is truncated at 70% for readability. The remaining H3K79 methylation after homozygous Dot1L deletion is probably due to the presence of some cells in which Cre is not expressed (yet). **P < 0.01 and ***P < 0.001 by two‐way ANOVA, comparison to wild type.
Representative H&E and immunohistochemical staining on sequential sections of thymic lymphomas of the indicated genotypes. A picture with lower magnification of independent samples is included in Fig EV3B.
Kaplan–Meier curves of mice harboring thymocytes with indicated genotypes. An event was defined as death or sacrifice of a mouse caused by a thymic lymphoma. Mice that died due to other causes or were still alive and event‐free at the end of the experiment were censored. Mice for which the cause of death could not be determined were removed from the data. Wild‐type mice were the Cre‐ littermates of the mice that were used for the other curves.
Summary of the data presented in panel C. A median latency could only be calculated when the tumor incidence was > 50%. The P value was determined by comparing to the Lck‐Cre;Hdac1 f/f curve with a Peto test, but a logrank test yielded the same conclusions.
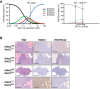
Yeast Dot1 is a distributive enzyme and shows waves of the different methylation states over a range of Dot1 concentrations in yeast (De Vos et al, 2011), Fig from De Vos et al (2017). The catalytic nature of mammalian DOT1L enzymes is not known, but the observation that the abundance of each methylation state increases upon Hdac1 deletion does not conflict with a distributive nature. Since H3K79 methylation levels are low in mammalian cells, it is possible we are looking at the start of the methylation waves, as indicated by the dotted lines.
Representative H&E and immunohistochemical staining of sequential sections of thymic lymphomas of the indicated genotypes. The scale bar represents 2 mm.
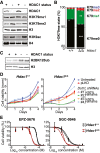
Immunoblots showing HDAC1 status and H3K79me/H3K9ac levels in nuclear lysates of Hdac1‐proficient (p53‐null) and Hdac1‐deficient thymic lymphoma cell lines. The top four and bottom two panels are from separate lysates of the same cell lines.
Mass spectrometry analysis of H3K79 methylation in the cell lines from panel A. Mean and individual data points of two independent cell lines; H3K79me0 is the predominant state, and the y‐axis is truncated at 80% for readability. **P < 0.01 and ***P < 0.001 by two‐way ANOVA.
Immunoblot showing H2BK120 ubiquitination levels in Hdac1‐proficient and Hdac1‐deficient cell lines (two independent lines each).
Growth curves of Hdac1‐proficient and Hdac1‐deficient cell lines that were left untreated or were infected with empty virus (pLKO) or shRNAs against Dot1L and selected with puromycin. Growth curves were determined by a series of resazurin assays, which measure metabolic activity, starting from four days post‐infection. Error bars indicate the range of two replicates from independent cell lines.
Inhibitor dose–response curves of the two DOT1L inhibitors EPZ‐5676 (Pinometostat) and SGC‐0946 in Hdac1‐proficient and Hdac1‐deficient cell lines. Cell viability was measured by a resazurin assay after three days of treatment, and measurements were normalized to DMSO‐treated cells. Two independent cell lines are plotted separately; error bars indicate the range of two biological replicates.

Knockdown efficiency of the shRNAs used in Fig 4D. The shRNAs 1 and 2 give a knockdown of more than 50%.
Immunoblot analysis of H3K79 methylation levels in the two cell lines with and without inhibitors. Both the effect of the inhibitors and the difference between the cell lines can be observed; * indicates a lane that was underloaded because most cells had died. For cells with EPZ‐5676 treatment, the H3K79me1 blot was re‐probed with α‐H3K79me2, following stripping the previous antibody and confirming that the stripping was complete. Finally, the blot was re‐probed with α‐H3‐C.

Representative apoptosis FACS plots of cell lines treated with DMSO or the DOT1L inhibitor SGC‐0946 for two days. Annexin V staining and DAPI staining were performed on unpermeabilized cells to distinguish live (Annexin V low; DAPI low), apoptotic (Annexin V high; DAPI low), and dead (Annexin V high; DAPI high) cells.
Quantification of several independent FACS experiments, including the experiment shown in panel (A). Mean with individual data points of 2–4 replicates each of two independent lines per genotype. ***P < 0.001 by two‐way ANOVA, comparison to corresponding DMSO control.
References
-
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR‐mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- NWO-VICI-016.130.627/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)/International
- NCI-KIEM-731.013.102/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)/International
- NCI-LIFT-731.015.405/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)/International
- Top 91213018/ZonMw (Netherlands Organisation for Health Research and Development)/International
- KWF2009-4511/KWF Kankerbestrijding (Dutch Cancer Society)/International
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

