In situ imaging of the bacterial flagellar motor disassembly and assembly processes
- PMID: 31304634
- PMCID: PMC6627242
- DOI: 10.15252/embj.2018100957
In situ imaging of the bacterial flagellar motor disassembly and assembly processes
Abstract
The self-assembly of cellular macromolecular machines such as the bacterial flagellar motor requires the spatio-temporal synchronization of gene expression with proper protein localization and association of dozens of protein components. In Salmonella and Escherichia coli, a sequential, outward assembly mechanism has been proposed for the flagellar motor starting from the inner membrane, with the addition of each new component stabilizing the previous one. However, very little is known about flagellar disassembly. Here, using electron cryo-tomography and sub-tomogram averaging of intact Legionella pneumophila, Pseudomonas aeruginosa, and Shewanella oneidensis cells, we study flagellar motor disassembly and assembly in situ. We first show that motor disassembly results in stable outer membrane-embedded sub-complexes. These sub-complexes consist of the periplasmic embellished P- and L-rings, and bend the membrane inward while it remains apparently sealed. Additionally, we also observe various intermediates of the assembly process including an inner-membrane sub-complex consisting of the C-ring, MS-ring, and export apparatus. Finally, we show that the L-ring is responsible for reshaping the outer membrane, a crucial step in the flagellar assembly process.
Keywords: assembly; bacterial flagellar motor; disassembly; electron cryo-tomography; in situ imaging.
© 2019 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A–O
(A, F, K) slices through electron cryo‐tomograms of L. pneumophila, P. aeruginosa, and S. oneidensis cells, respectively, highlighting a PL sub‐complex in the outer membrane (dashed yellow circle). (B, G, L) Enlarged views of the sub‐complexes. (C, H, M) Sub‐tomogram averages of PL sub‐complexes from each species. (D, I, N) Schematic representations of the sub‐tomogram averages with different rings colored and labeled. (E, J, O) Sub‐tomogram averages of fully assembled flagella from each species for comparison. Scale bars: (black) 50 nm, (orange) 25 nm, and (red) 20 nm.

- A
Sub‐tomogram average of the IM sub‐complex constituting the C‐ ring, MS‐ring, and export apparatus.
- B
Schematic representation of the sub‐tomogram average shown in (A) highlighting the different parts of the complex.
- C
Sub‐tomogram average of the motors of fully assembled flagella.
- D–J
Slices through electron cryo‐tomograms showing neighboring PL and IM sub‐complexes. Dashed yellow circles highlight the IM sub‐complex, while dashed blue arrows highlight the PL sub‐complex. Dashed red lines mark the border between two images used to make a composite image when the PL and IM sub‐complexes were found at different levels in the tomogram.
- K
Schematic representation of an IM sub‐complex in the vicinity of a PL sub‐complex.
- L, M
Slices through electron cryo‐tomograms of L. pneumophila highlighting the presence of a fully assembled basal body lacking the hook and the filament.
- N
Schematic representation of the basal bodies in (L and M).
- O
Central slice through an electron cryo‐tomogram of a lysed cell. The dashed yellow circle highlights the flagellar motor.
- P
Enlarged view of the same slice shown in (O). The absence of the C‐ring and the export apparatus is highlighted by the dashed orange arrow.
- Q
Schematic representation of the complex found in (P).

- A–C
Slices through electron cryo‐tomograms showing fully assembled motors without the hook and filament. The dashed yellow circles indicate the IM sub‐complex.
- D
Schematic representation of the P. aeruginosa motors lacking the hook and filament shown in (A–C).
- E–G
Slices through electron cryo‐tomograms showing fully assembled motors with the hook and lacking the filament.
- H
Schematic representation of the motors with the hook shown in (E–G).
- I–K
Slices through electron cryo‐tomograms of intact P. aeruginosa ΔflgI cells showing the presence of the inner‐membrane sub‐complex with the rod.
- L
Schematic representation of the inner‐membrane sub‐complex with the rod shown in (I‐K).
- M–O
Slices through electron cryo‐tomograms of lysed P. aeruginosa ΔflgI cells showing the presence of the sub‐complex constituting the MS‐ring and the rod.
- P
Schematic representation of the sub‐complex described in (M–O).
- Q
A slice through an electron cryo‐tomogram of a P. aeruginosa ΔflgG cell highlighting the presence of the inner‐membrane sub‐complex.
- R
Schematic representation of the inner‐membrane sub‐complex shown in (Q).

- A–C
Slices through electron cryo‐tomograms of wild‐type cells showing fully assembled motors without the hook and filament.
- D
Schematic representation of the motors lacking the hook and filament shown in (A–C).
- E
Sub‐tomogram average of the motors of fully assembled flagella.
- F
Slice through an electron cryo‐tomogram of a ΔflgH cell showing an IM sub‐complex, indicated by the dashed yellow circle.
- G
Schematic representation of the IM sub‐complex shown in (F).
- H, I
Slices through electron cryo‐tomograms of ΔflgH cells showing the IM sub‐complex with the rod and the P‐ring, indicated by the dashed yellow circles.
- J
Schematic representation of the structures shown in (H and I).
- K–M
Slices through electron cryo‐tomograms of ΔflaA/B cells highlighting the flagellar motor and the hook (without the filament).
- N
Sub‐tomogram average of the flagellar motor and the hook structure found in ΔflaA/B cells.
- O
Schematic representation of the sub‐tomogram average shown in (N).
- P–R
Slices through electron cryo‐tomograms of ΔflaA/B cells illustrating a disassembly product constituting the PL sub‐complex, the rod, and the hook.
- S
Sub‐tomogram average of the disassembly complex shown in (P–R). The dashed orange arrow indicates the absence of the IM sub‐complex in this structure.
- T
Schematic representation of the disassembly product shown in (P–S).
- U–W
Slices through electron cryo‐tomograms of ΔflaA/B cells highlighting PL sub‐complexes (dashed yellow circles).
- X
Sub‐tomogram average of the PL sub‐complexes in ΔflaA/B cells.
- Y
Schematic representation of the sub‐tomogram average shown in (X).
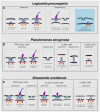
- A–C
Schematic representations of the various assembly and disassembly sub‐complexes observed in this study in L. pneumophila, P. aeruginosa, and S. oneidensis (wild‐type and mutant strains), respectively. The blue shaded area in (A) represents an intact IM sub‐complex in the vicinity of a PL sub‐complex. Note that the boundaries between the different parts in these schematics are tentative.
Similar articles
-
In Situ Structures of Polar and Lateral Flagella Revealed by Cryo-Electron Tomography.J Bacteriol. 2019 Jun 10;201(13):e00117-19. doi: 10.1128/JB.00117-19. Print 2019 Jul 1. J Bacteriol. 2019. PMID: 31010901 Free PMC article.
-
Bacterial flagellar motor PL-ring disassembly subcomplexes are widespread and ancient.Proc Natl Acad Sci U S A. 2020 Apr 21;117(16):8941-8947. doi: 10.1073/pnas.1916935117. Epub 2020 Apr 2. Proc Natl Acad Sci U S A. 2020. PMID: 32241888 Free PMC article.
-
Novel transient cytoplasmic rings stabilize assembling bacterial flagellar motors.EMBO J. 2022 May 16;41(10):e109523. doi: 10.15252/embj.2021109523. Epub 2022 Mar 18. EMBO J. 2022. PMID: 35301732 Free PMC article.
-
Molecular architecture of the bacterial flagellar motor in cells.Biochemistry. 2014 Jul 15;53(27):4323-33. doi: 10.1021/bi500059y. Epub 2014 Jul 1. Biochemistry. 2014. PMID: 24697492 Free PMC article. Review.
-
Undiscovered regions on the molecular landscape of flagellar assembly.Curr Opin Microbiol. 2015 Dec;28:98-105. doi: 10.1016/j.mib.2015.08.011. Epub 2015 Oct 23. Curr Opin Microbiol. 2015. PMID: 26490009 Review.
Cited by
-
Construction and Loss of Bacterial Flagellar Filaments.Biomolecules. 2020 Nov 9;10(11):1528. doi: 10.3390/biom10111528. Biomolecules. 2020. PMID: 33182435 Free PMC article. Review.
-
Structural Conservation and Adaptation of the Bacterial Flagella Motor.Biomolecules. 2020 Oct 29;10(11):1492. doi: 10.3390/biom10111492. Biomolecules. 2020. PMID: 33138111 Free PMC article. Review.
-
Mechanisms of E. coli chemotaxis signaling pathways visualized using cryoET and computational approaches.Biochem Soc Trans. 2022 Dec 16;50(6):1595-1605. doi: 10.1042/BST20220191. Biochem Soc Trans. 2022. PMID: 36421737 Free PMC article. Review.
-
A Tripartite Efflux System Affects Flagellum Stability in Helicobacter pylori.Int J Mol Sci. 2022 Oct 1;23(19):11609. doi: 10.3390/ijms231911609. Int J Mol Sci. 2022. PMID: 36232924 Free PMC article.
-
Flagella, Chemotaxis and Surface Sensing.Adv Exp Med Biol. 2022;1386:185-221. doi: 10.1007/978-3-031-08491-1_7. Adv Exp Med Biol. 2022. PMID: 36258073
References
-
- Agulleiro JI, Fernandez JJ (2011) Fast tomographic reconstruction on multicore computers. Bioinformatics 27: 582–583 - PubMed
-
- Aldridge P, Jenal U (1999) Cell cycle‐dependent degradation of a flagellar motor component requires a novel‐type response regulator. Mol Microbiol 32: 379–391 - PubMed
-
- Baker LA, Folkers GE, Sinnige T, Houben K, Kaplan M, van der Cruijsen EAW, Baldus M (2015) Magic‐angle‐spinning solid‐state NMR of membrane proteins In Methods in enzymology, Shukla AK. (ed.), pp 307–328. Elsevier; - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

