Comparative genomics identifies potential virulence factors in Clostridium tertium and C. paraputrificum
- PMID: 31304854
- PMCID: PMC6629180
- DOI: 10.1080/21505594.2019.1637699
Comparative genomics identifies potential virulence factors in Clostridium tertium and C. paraputrificum
Abstract
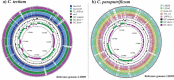
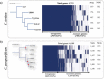
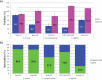
Some well-known Clostridiales species such as Clostridium difficile and C. perfringens are agents of high impact diseases worldwide. Nevertheless, other foreseen Clostridiales species have recently emerged such as Clostridium tertium and C. paraputrificum. Three fecal isolates were identified as Clostridium tertium (Gcol.A2 and Gcol.A43) and C. paraputrificum (Gcol.A11) during public health screening for C. difficile infections in Colombia. C. paraputrificum genomes were highly diverse and contained large numbers of accessory genes. Genetic diversity and accessory gene percentage were lower among the C. tertium genomes than in the C. paraputrificum genomes. C. difficile tcdA and tcdB toxins encoding homologous sequences and other potential virulence factors were also identified. EndoA interferase, a toxic component of the type II toxin-antitoxin system, was found among the C. tertium genomes. toxA was the only toxin encoding gene detected in Gcol.A43, the Colombian isolate with an experimentally-determined high cytotoxic effect. Gcol.A2 and Gcol.A43 had higher sporulation efficiencies than Gcol.A11 (84.5%, 83.8% and 57.0%, respectively), as supported by the greater number of proteins associated with sporulation pathways in the C. tertium genomes compared with the C. paraputrificum genomes (33.3 and 28.4 on average, respectively). This work allowed complete genome description of two clostridiales species revealing high levels of intra-taxa diversity, accessory genomes containing virulence-factors encoding genes (especially in C. paraputrificum), with proteins involved in sporulation processes more highly represented in C. tertium. These finding suggest the need to advance in the study of those species with potential importance at public health level.
Keywords: Clostridial species; Clostridium paraputrificum; Clostridium tertium; genetic diversity; virulence factors.
Figures

Similar articles
-
Preterm Infant-Associated Clostridium tertium, Clostridium cadaveris, and Clostridium paraputrificum Strains: Genomic and Evolutionary Insights.Genome Biol Evol. 2017 Oct 1;9(10):2707-2714. doi: 10.1093/gbe/evx210. Genome Biol Evol. 2017. PMID: 29044436 Free PMC article.
-
Genomic and phenotypic studies among Clostridioides difficile isolates show a high prevalence of clade 2 and great diversity in clinical isolates from Mexican adults and children with healthcare-associated diarrhea.Microbiol Spectr. 2024 Jul 2;12(7):e0394723. doi: 10.1128/spectrum.03947-23. Epub 2024 Jun 12. Microbiol Spectr. 2024. PMID: 38864670 Free PMC article.
-
Global insights into the genome dynamics of Clostridioides difficile associated with antimicrobial resistance, virulence, and genomic adaptations among clonal lineages.Front Cell Infect Microbiol. 2025 Jan 15;14:1493225. doi: 10.3389/fcimb.2024.1493225. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39882343 Free PMC article.
-
Hype or hypervirulence: a reflection on problematic C. difficile strains.Virulence. 2013 Oct 1;4(7):592-6. doi: 10.4161/viru.26297. Epub 2013 Sep 10. Virulence. 2013. PMID: 24060961 Free PMC article. Review.
-
Large Clostridial Toxins: Mechanisms and Roles in Disease.Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0006421. doi: 10.1128/MMBR.00064-21. Epub 2021 Jun 2. Microbiol Mol Biol Rev. 2021. PMID: 34076506 Free PMC article. Review.
Cited by
-
Whole genome sequence of bacteremic Clostridium tertium in a World War I soldier, 1914.Curr Res Microb Sci. 2021 Dec 4;3:100089. doi: 10.1016/j.crmicr.2021.100089. eCollection 2022. Curr Res Microb Sci. 2021. PMID: 34984406 Free PMC article.
-
Intra-species diversity of Clostridium perfringens: A diverse genetic repertoire reveals its pathogenic potential.Front Microbiol. 2022 Jul 22;13:952081. doi: 10.3389/fmicb.2022.952081. eCollection 2022. Front Microbiol. 2022. PMID: 35935202 Free PMC article.
-
Completion of the gut microbial epi-bile acid pathway.Gut Microbes. 2021 Jan-Dec;13(1):1-20. doi: 10.1080/19490976.2021.1907271. Gut Microbes. 2021. PMID: 33938389 Free PMC article.
-
Comparative Genomics of Clostridium baratii Reveals Strain-Level Diversity in Toxin Abundance.Microorganisms. 2022 Jan 20;10(2):213. doi: 10.3390/microorganisms10020213. Microorganisms. 2022. PMID: 35208668 Free PMC article.
-
Identification of mucin degraders of the human gut microbiota.Sci Rep. 2021 May 27;11(1):11094. doi: 10.1038/s41598-021-90553-4. Sci Rep. 2021. PMID: 34045537 Free PMC article.
References
-
- Tracy BP, Jones SW, Fast AG, et al. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol. 2012;23:364–381. - PubMed
-
- Kunisawa T. Evolutionary relationships of completely sequenced Clostridia species and close relatives. Int J Syst Evol Microbiol. 2015;65:4276–4283. - PubMed
-
- Rainey FA. Clostridiales. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd; 2015. Available from: https://onlinelibrary.wiley.com/doi/book/10.1002/9781118960608 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
