Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain
- PMID: 31304985
- PMCID: PMC6717894
- DOI: 10.15252/embj.2018100481
Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain
Abstract
Regulation of adult neural stem cell (NSC) number is critical for lifelong neurogenesis. Here, we identified a post-transcriptional control mechanism, centered around the microRNA 204 (miR-204), to control the maintenance of quiescent (q)NSCs. miR-204 regulates a spectrum of transcripts involved in cell cycle regulation, neuronal migration, and differentiation in qNSCs. Importantly, inhibition of miR-204 function reduced the number of qNSCs in the subependymal zone (SEZ) by inducing pre-mature activation and differentiation of NSCs without changing their neurogenic potential. Strikingly, we identified the choroid plexus of the mouse lateral ventricle as the major source of miR-204 that is released into the cerebrospinal fluid to control number of NSCs within the SEZ. Taken together, our results describe a novel mechanism to maintain adult somatic stem cells by a niche-specific miRNA repressing activation and differentiation of stem cells.
Keywords: adult neurogenesis; miR-204; neural stem cells; neurogenesis; neurogenic priming.
© 2019 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
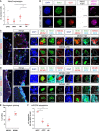
- A
Dot plot depicting the expression of mRNA for neurogenic transcription factor MEIS2 in prospectively isolated cellular populations from the SEZ.
- B
Micrographs depicting the expression of neurogenic fate determinant Meis2 mRNA and MEIS2 protein in acutely dissociated SEZ cells. Note that Meis2 mRNA‐positive LRCs have low (no) MEIS2 protein.
- C, D
IHC labeling for MEIS2 (C) and MCM6 (D) of LRCs positive for BrdU‐only and neuroblasts marked by DCX.
- E
Dot plot showing the proportion of BrdU+ LRCs negative for MEIS2 or MCM6 protein.
- F
Dot plot depicting the expression of miR‐204 in prospectively isolated cells of neural lineage.

Dot plots depicting the expression of mRNA for neurogenic transcription factors Sox11, Arx, Dlx1, and Dlx2 in prospectively isolated cellular populations from the SEZ.
Venn diagram depicting common neural stem enriched genes from three different publications.
Graph showing GO terms enriched in the set of 74 overlapping genes identified in (B). GO terms related to neurogenesis are shown in red and to the cell cycle in magenta.
Micrographs showing neural stem cells (hGFAP‐GFP+; CD133+) isolated from the BrdU‐treated hGFAP‐eGFP animals and fixed 2 h after plating.
Table showing predicted microRNA‐mRNA interactions and functional regulations.
Pie chart depicting predicted and published targets of miR‐204 among priming factors following the expression pattern DE < qNSC < aNSC < NB.
Dot plot depicting the functional regulation of neurogenic priming factors by miR‐204 in luciferase reporter assays.
Graph showing GO terms enriched in the set of 34 putative miR‐204 targets. GO terms related to neurogenesis are shown in red and to the cell cycle in magenta.
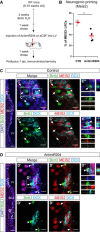
- A
Schematic representation of the experimental workflow for labeling LRCs with BrdU in the adult SEZ and miR‐204 inhibition using antagomirs.
- B
Dot plot depicting the immunoreactivity of LRCs for MEIS2 in control and antagomirs treated animals 7 dpi.
- C, D
Micrographs showing IHC for the neurogenic priming factor MEIS2 in the SEZ 7 days after injection of artificial CSF (control, C) or AntimiR204 (D). White arrows indicate LRCs without detectable MEIS2 protein. The white arrowheads are pointing out the loss of neurogenic priming (LRCs positive for MEIS2 protein) upon the antagomir treatments.
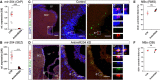
- A, B
Dot plots showing miR‐204 levels in ChP (A) and in the SEZ (B) 3 days after injection of aCSF or AntimiR204 into the lateral ventricle.
- C, D
Micrographs depicting the TUNEL staining in the SEZ 7 days after injection of aCSF (control, C) or AntimiR204 (D). Boxed areas correspond to the higher magnifications in the adjacent panels (C‘, C‘‘, D‘, D‘‘). Note that almost no BrdU+ TUNEL+ cells could be found in any of the analyzed conditions.
- E, F
Dot plots depicting the proportion of the neuroblasts (BrdU+ DCX+) from label‐retaining cells in the RMS (E) and in the OB (F) 7 days after aCSF or AntimiR204 injection.
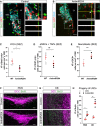
- A, B
Micrographs depicting the cellular composition in the SEZ 7 days after injection of aCSF (control, A) or AntimiR204 (B). Boxed areas correspond to higher magnifications in adjacent panels (A‘, B‘).
- C–E
Dot plots depicting the abundance of LRCs (BrdU+, Ki67−, DCX−), aNSCs and TAPs (BrdU−, Ki67+, DCX−) and neuroblasts (DCX+ Ki67+ or DCX+ Ki67−) in the SEZ 7 days after aCSF or AntimiR204 injection. Note that the number of LRCs (white arrows in A and B) is reduced in Antimir204 injected SEZ, while the number of NBs is increased.
- F, G
Micrographs depicting the elbow—the most ventral region of the RMS (F) and OB neuronal layers (G) upon injection of artificial CSF (control) or AntimiR204.
- H
Dot plot showing the density of BrdU+ cells (progeny of NSCs) in the RMS and in the OB 7 days after artificial CSF or AntimiR204 injection.

- A
Volcano plot illustrating regulated transcripts in qNSC 3 days after AntimiR204 injection in the lateral ventricle (fold change > 2 and q‐value < 0.05 are depicted with red line).
- B
Dot plot depicting levels of neurogenic fate determinants targeted by miR‐204 in control and miR‐204‐deficient qNSCs. Data are shown as mean ± SEM; each symbol represents independent biological replicate.
- C, D
Histogram showing GO terms biological processes (C) and cellular compartments (D) enriched (fold enrichment > 2, P‐values are indicated on the bars) in the set of genes up‐regulated after miR‐204 inhibition as shown in (A).
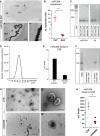
- A
Micrographs of in situ hybridization for miR‐204 in an adult mouse brain section. (A‘ and A‘‘) are magnifications of boxed areas in (A or A‘), respectively.
- B
Dot plot showing miR‐204 levels in ChP and SEZ in the adult mouse brain measured by RT–qPCR.
- C
Agarose gel of RT–qPCR product loaded after the saturation phase showing presence of miR‐204 and U6 in mouse ChP, SEZ, and CSF.
- D, E
Plots depicting the number and size of EVs isolated from human CSF (D) and miR‐204 levels (E) in EVs and EV‐free CSF (n = 2).
- F
Agarose gel of RT–qPCR analysis depicting levels of miR‐204 in the EV‐free supernatant and EV containing fraction of human CSF loaded at the saturation phase.
- G
Micrographs of transmission electron microscopy imaged extracellular vesicles (EVs) isolated from sham‐treated (upper row) and GW4869‐treated animals (lower row). (G‘ and G‘‘) are magnifications of boxed areas in overview images to the left.
- H
Dot plot depicting miR‐204 levels in the CSF isolated 2 h after the ventricular injection of GW4869 inhibitor.
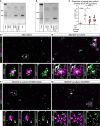
- A
Gel pictures of the saturation phase of RT–qPCR analysis for miR‐204 in SEZ primary culture cells, ChP co‐culture, and ChP explants 7 day post‐preparation.
- B
Gel electrophoresis of RT–qPCR analysis loaded at the saturation phase for miR‐204 in SEZ culture medium and SEZ medium from the ChP co‐culture.
- C
Dot plot showing the proportion of neural stem cells (GFAP+) out of DAPI+ cells in primary SEZ/ChP co‐culture with or without treatment with GW4869 inhibitor or AntimiR204.
- D–G
Micrographs depicting the cellular compositions of SEZ cultures after co‐culturing with control‐treated, GW4869 inhibitor‐treated, or AntimiR204‐treated ChP 7 days after plating. Boxed areas correspond to the higher magnification images in adjacent panels (D‘, E‘, F‘, G‘).
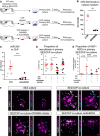
Schematic representation of the experimental setup for primary SEZ/ChP co‐culture.
Dot plot representing RT–qPCR analysis of the miR‐204 levels in the medium after 24‐h incubation of ChP with GW4869 inhibitor or DMSO control.
Dot plot showing miR‐204 levels in the ChP used for the co‐culture 1 day after incubation with or without AntimiR204.
Dot plot showing the proportion of DCX‐positive cells out of DAPI+ cells in primary SEZ/ChP co‐culture without treatment or treated with GW4869 inhibitor or AntimiR204.
Dot plot depicting the proportion of proliferative (Ki67+) neural stem cells out of DAPI+ cells in primary SEZ/ChP co‐culture without treatment or treated with GW4869 inhibitor or AntimiR204.
Micrographs showing Ki67+ GFAP+ cells within SEZ cultures after co‐culturing with control‐treated, GW4869 inhibitor‐treated, or AntimiR204‐treated ChP 7 days after plating.
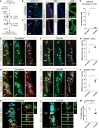
- A
Schematic representation of the experimental setup to address the effect of ChP‐specific miR‐204 inhibition on the number of LRCs and neurogenic priming.
- B, C
Micrographs depicting specific expression of AAV5 encoded GFP in the ChP 7 days after viral delivery in the lateral ventricle. (B‘ and C‘) are magnifications of the SEZ and (B‘‘ and C‘‘) of the ChP.
- D
Dot plot depicting expression of miR‐204 in ChP 7 days after second ventricular injection of AAV5 encoding for miR‐204‐specific TuD compared to scrambled control.
- E–J
Micrographs depicting immunoreactivity for MEIS2 (E, F) and MCM6 (H, I) priming factors in the LRCs (BrdU+ only) 7 days after ChP‐specific miR‐204 inhibition (F, I) and scrambled control (E, H). White arrows point out the primed LRCs, the arrowheads label the MEIS2+ or MCM6+ + LRCs. (G, J) Dot plots depicting the proportion of LRCs immunoreactive for the priming proteins MEIS2 (G) and MCM6 (J) after ChP‐specific miR‐204 interference.
- K, L
Micrographs showing the cellular composition in the SEZ 7 days after injection of AVV5 encoding for scrambled control (K) and miR‐204‐specific TuD (L). Note the reduction of primed LRCs (BrdU+ only, white arrows in K and L) upon TuD‐204 injection.
- M
Dot plot depicting the number of LRCs (BrdU+ only) in the SEZ 7 days after miR‐204 inhibition.

Scheme showing the constructs for AAVs serotype 5 encoding for either a scrambled control or miR‐204 specific tough decoys (TuD‐204).
Schematic representation of the experimental setup to address the specificity of the AAVs serotype 5 and the effect of loss of miR‐204 function in the ChP.
FACS analysis of the cells isolated from the SEZ 7 dpi after injection of AAV5 encoding for scrambled control and miR‐204‐specific TuD. Note that almost no GFP+ cells could be detected.
Dot plot depicting the levels of miR‐204 in ChP 7 days after second ventricular injection of AAV5 encoding for miR‐204‐specific TuD compared to scrambled control. Same symbols represent ChP isolated from left and right hemisphere.
Volcano plot illustrating regulated transcripts after TuD‐204 injection in the ChP (fold change > 2 and q‐value < 0.05 are depicted with red line).
Venn diagram depicting overlap of regulated genes shown in (E) and predicted miR‐204 targets.
Histogram showing KEGG pathways enrichment (fold enrichment > 2, P‐values are indicated on the bars) of the regulated genes shown in (F).
Comment in
-
Sleep or deplete: how the choroid plexus helps to keep neural stem cells in balance.EMBO J. 2019 Sep 2;38(17):e103013. doi: 10.15252/embj.2019103013. Epub 2019 Aug 21. EMBO J. 2019. PMID: 31432524 Free PMC article.
References
-
- Agoston Z, Heine P, Brill MS, Grebbin BM, Hau AC, Kallenborn‐Gerhardt W, Schramm J, Gotz M, Schulte D (2014) Meis2 is a Pax6 co‐factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141: 28–38 - PubMed
-
- Azim K, Fischer B, Hurtado‐Chong A, Draganova K, Cantu C, Zemke M, Sommer L, Butt A, Raineteau O (2014) Persistent Wnt/beta‐catenin signaling determines dorsalization of the postnatal subventricular zone and neural stem cell specification into oligodendrocytes and glutamatergic neurons. Stem Cells 32: 1301–1312 - PubMed
-
- Basak O, Krieger TG, Muraro MJ, Wiebrands K, Stange DE, Frias‐Aldeguer J, Rivron NC, van de Wetering M, van Es JH, van Oudenaarden A et al (2018) Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proc Natl Acad Sci USA 115: E610–E619 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- SPP1738/Deutsche Forschungsgemeinschaft (DFG)/International
- SPP1757/Deutsche Forschungsgemeinschaft (DFG)/International
- SFB870/Deutsche Forschungsgemeinschaft (DFG)/International
- HU1961/2-1/Deutsche Forschungsgemeinschaft (DFG)/International
- SCHU1218/3-3/Deutsche Forschungsgemeinschaft (DFG)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

