The cysteine-free single mutant C32S of APEX2 is a highly expressed and active fusion tag for proximity labeling applications
- PMID: 31306516
- PMCID: PMC6699085
- DOI: 10.1002/pro.3685
The cysteine-free single mutant C32S of APEX2 is a highly expressed and active fusion tag for proximity labeling applications
Abstract
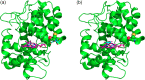
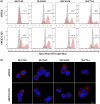
APEX2, an engineered ascorbate peroxidase for high activity, is a powerful tool for proximity labeling applications. Owing to its lack of disulfides and the calcium-independent activity, APEX2 can be applied intracellularly for targeted electron microscopy imaging or interactome mapping when fusing to a protein of interest. However, APEX2 fusion is often deleterious to the protein expression, which seriously hampers its wide utility. This problem is especially compelling when APEX2 is fused to structurally delicate proteins, such as multi-pass membrane proteins. In this study, we found that a cysteine-free single mutant C32S of APEX2 dramatically improved the expression of fusion proteins in mammalian cells without compromising the enzyme activity. We fused APEX2 and APEX2C32S to four multi-transmembrane solute carriers (SLCs), SLC1A5, SLC6A5, SLC6A14, and SLC7A1, and compared their expressions in stable HEK293T cell lines. Except the SLC6A5 fusions expressing at decent levels for both APEX2 (70%) and APEX2C32S (73%), other three SLC proteins showed significantly better expression when fusing to APEX2C32S (69 ± 13%) than APEX2 (29 ± 15%). Immunofluorescence and western blot experiments showed correct plasma membrane localization and strong proximity labeling efficiency in all four SLC-APEX2C32S cells. Enzyme kinetic experiments revealed that APEX2 and APEX2C32S have comparable activities in terms of oxidizing guaiacol. Overall, we believe APEX2C32S is a superior fusion tag to APEX2 for proximity labeling applications, especially when mismatched disulfide bonding or poor expression is a concern.
Keywords: APEX2; ascorbate peroxidase; proximity labeling; solute carriers.
© 2019 The Protein Society.
Figures




Similar articles
-
Expanding APEX2 Substrates for Proximity-Dependent Labeling of Nucleic Acids and Proteins in Living Cells.Angew Chem Int Ed Engl. 2019 Aug 19;58(34):11763-11767. doi: 10.1002/anie.201905949. Epub 2019 Jul 26. Angew Chem Int Ed Engl. 2019. PMID: 31240809
-
Optimized APEX2 peroxidase-mediated proximity labeling in fast- and slow-growing mycobacteria.Methods Enzymol. 2022;664:267-289. doi: 10.1016/bs.mie.2021.11.021. Epub 2021 Dec 30. Methods Enzymol. 2022. PMID: 35331378 Free PMC article.
-
APEX2 Proximity Proteomics Resolves Flagellum Subdomains and Identifies Flagellum Tip-Specific Proteins in Trypanosoma brucei.mSphere. 2021 Feb 10;6(1):e01090-20. doi: 10.1128/mSphere.01090-20. mSphere. 2021. PMID: 33568455 Free PMC article.
-
Solute carriers: The gatekeepers of metabolism.Cell. 2025 Feb 20;188(4):869-884. doi: 10.1016/j.cell.2025.01.015. Cell. 2025. PMID: 39983672 Review.
-
Enzyme Fusions in Biocatalysis: Coupling Reactions by Pairing Enzymes.Chembiochem. 2019 Jan 2;20(1):20-28. doi: 10.1002/cbic.201800394. Epub 2018 Oct 2. Chembiochem. 2019. PMID: 30178909 Free PMC article. Review.
Cited by
-
Advances in enzyme-mediated proximity labeling and its potential for plant research.Plant Physiol. 2022 Feb 4;188(2):756-768. doi: 10.1093/plphys/kiab479. Plant Physiol. 2022. PMID: 34662401 Free PMC article.
-
Next-Generation Protein-Ligand Interaction Networks: APEX as a Powerful Technology.Proteomes. 2025 Jun 23;13(3):26. doi: 10.3390/proteomes13030026. Proteomes. 2025. PMID: 40700270 Free PMC article. Review.
-
Recent progress in mass spectrometry-based strategies for elucidating protein-protein interactions.Cell Mol Life Sci. 2021 Jul;78(13):5325-5339. doi: 10.1007/s00018-021-03856-0. Epub 2021 May 27. Cell Mol Life Sci. 2021. PMID: 34046695 Free PMC article. Review.
-
Proximity Protein Labeling In Dictyostelium With Engineered Ascorbic Acid Peroxidase 2.J Biol Methods. 2023 Mar 16;10:e99010002. doi: 10.14440/jbm.2023.396. eCollection 2023. J Biol Methods. 2023. PMID: 37007980 Free PMC article.
-
Proximity-dependent labeling methods for proteomic profiling in living cells: An update.Wiley Interdiscip Rev Dev Biol. 2021 Jan;10(1):e392. doi: 10.1002/wdev.392. Epub 2020 Sep 10. Wiley Interdiscip Rev Dev Biol. 2021. PMID: 32909689 Free PMC article. Review.
References
-
- Buchwalow I, Boecker W, Wolf E, Samoilova V, Tiemann M. Signal amplification in immunohistochemistry: loose‐jointed deformable heteropolymeric HRP conjugates vs. linear polymer backbone HRP conjugates. Acta Histochem. 2013;115:587–594. - PubMed
-
- Toda Y, Kono K, Abiru H, et al. Application of tyramide signal amplification system to immunohistochemistry: a potent method to localize antigens that are not detectable by ordinary method. Pathol Int. 1999;49:479–483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

