Muscle xenografts reproduce key molecular features of facioscapulohumeral muscular dystrophy
- PMID: 31306642
- PMCID: PMC6730665
- DOI: 10.1016/j.expneurol.2019.113011
Muscle xenografts reproduce key molecular features of facioscapulohumeral muscular dystrophy
Abstract
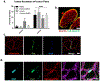
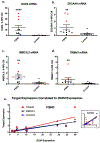
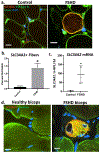
Aberrant expression of DUX4, a gene unique to humans and primates, causes Facioscapulohumeral Muscular Dystrophy-1 (FSHD), yet the pathogenic mechanism is unknown. As transgenic overexpression models have largely failed to replicate the genetic changes seen in FSHD, many studies of endogenously expressed DUX4 have been limited to patient biopsies and myogenic cell cultures, which never fully differentiate into mature muscle fibers. We have developed a method to xenograft immortalized human muscle precursor cells from patients with FSHD and first-degree relative controls into the tibialis anterior muscle compartment of immunodeficient mice, generating human muscle xenografts. We report that FSHD cells mature into organized and innervated human muscle fibers with minimal contamination of murine myonuclei. They also reconstitute the satellite cell niche within the xenografts. FSHD xenografts express DUX4 and DUX4 downstream targets, retain the 4q35 epigenetic signature of their original donors, and express a novel protein biomarker of FSHD, SLC34A2. Ours is the first scalable, mature in vivo human model of FSHD. It should be useful for studies of the pathogenic mechanism of the disease as well as for testing therapeutic strategies targeting DUX4 expression.
Keywords: Biomarkers; DUX4; FSHD; Facioscapulohumeral muscular dystrophy; Hypomethylation; Satellite cells; Xenograft; hMPCs.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


Comment in
-
A Comment on "Muscle Xenografts Reproduce Key Molecular Features of Facioscapulohumeral Muscular Dystrophy": What Is New and What Has Already been Done and Reported but Was Not Quoted?Cell Transplant. 2020 Jan-Dec;29:963689720939120. doi: 10.1177/0963689720939120. Cell Transplant. 2020. PMID: 32830546 Free PMC article.
References
-
- van Deutekom JC, et al. (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Human molecular genetics 2(12):2037–2042. - PubMed
-
- Lemmers RJ, et al. (2002) Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nature genetics 32(2):235–236. - PubMed
-
- Gabriels J, et al. (1999) Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236(1):25–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

