PhenPath: a tool for characterizing biological functions underlying different phenotypes
- PMID: 31307376
- PMCID: PMC6631446
- DOI: 10.1186/s12864-019-5868-x
PhenPath: a tool for characterizing biological functions underlying different phenotypes
Abstract
Background: Many diseases are associated with complex patterns of symptoms and phenotypic manifestations. Parsimonious explanations aim at reconciling the multiplicity of phenotypic traits with the perturbation of one or few biological functions. For this, it is necessary to characterize human phenotypes at the molecular and functional levels, by exploiting gene annotations and known relations among genes, diseases and phenotypes. This characterization makes it possible to implement tools for retrieving functions shared among phenotypes, co-occurring in the same patient and facilitating the formulation of hypotheses about the molecular causes of the disease.
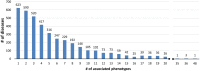
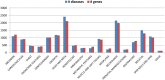
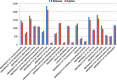
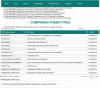
Results: We introduce PhenPath, a new resource consisting of two parts: PhenPathDB and PhenPathTOOL. The former is a database collecting the human genes associated with the phenotypes described in Human Phenotype Ontology (HPO) and OMIM Clinical Synopses. Phenotypes are then associated with biological functions and pathways by means of NET-GE, a network-based method for functional enrichment of sets of genes. The present version considers only phenotypes related to diseases. PhenPathDB collects information for 18 OMIM Clinical synopses and 7137 HPO phenotypes, related to 4292 diseases and 3446 genes. Enrichment of Gene Ontology annotations endows some 87.7, 86.9 and 73.6% of HPO phenotypes with Biological Process, Molecular Function and Cellular Component terms, respectively. Furthermore, 58.8 and 77.8% of HPO phenotypes are also enriched for KEGG and Reactome pathways, respectively. Based on PhenPathDB, PhenPathTOOL analyzes user-defined sets of phenotypes retrieving diseases, genes and functional terms which they share. This information can provide clues for interpreting the co-occurrence of phenotypes in a patient.
Conclusions: The resource allows finding molecular features useful to investigate diseases characterized by multiple phenotypes, and by this, it can help researchers and physicians in identifying molecular mechanisms and biological functions underlying the concomitant manifestation of phenotypes. The resource is freely available at http://phenpath.biocomp.unibo.it .
Keywords: Biological process; Diseases; Enrichment; Molecular pathway; Phenotype.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
eDGAR: a database of Disease-Gene Associations with annotated Relationships among genes.BMC Genomics. 2017 Aug 11;18(Suppl 5):554. doi: 10.1186/s12864-017-3911-3. BMC Genomics. 2017. PMID: 28812536 Free PMC article.
-
NET-GE: a novel NETwork-based Gene Enrichment for detecting biological processes associated to Mendelian diseases.BMC Genomics. 2015;16 Suppl 8(Suppl 8):S6. doi: 10.1186/1471-2164-16-S8-S6. Epub 2015 Jun 18. BMC Genomics. 2015. PMID: 26110971 Free PMC article.
-
HPOSim: an R package for phenotypic similarity measure and enrichment analysis based on the human phenotype ontology.PLoS One. 2015 Feb 9;10(2):e0115692. doi: 10.1371/journal.pone.0115692. eCollection 2015. PLoS One. 2015. PMID: 25664462 Free PMC article.
-
The human phenotype ontology.Clin Genet. 2010 Jun;77(6):525-34. doi: 10.1111/j.1399-0004.2010.01436.x. Epub 2010 Feb 11. Clin Genet. 2010. PMID: 20412080 Review.
-
Curation and expansion of Human Phenotype Ontology for defined groups of inborn errors of immunity.J Allergy Clin Immunol. 2022 Jan;149(1):369-378. doi: 10.1016/j.jaci.2021.04.033. Epub 2021 May 12. J Allergy Clin Immunol. 2022. PMID: 33991581 Free PMC article.
Cited by
-
Mapping OMIM Disease-Related Variations on Protein Domains Reveals an Association Among Variation Type, Pfam Models, and Disease Classes.Front Mol Biosci. 2021 May 7;8:617016. doi: 10.3389/fmolb.2021.617016. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026820 Free PMC article.
-
VarI-COSI 2018: a forum for research advances in variant interpretation and diagnostics.BMC Genomics. 2019 Jul 16;20(Suppl 8):550. doi: 10.1186/s12864-019-5862-3. BMC Genomics. 2019. PMID: 31307380 Free PMC article. No abstract available.
-
Rare deleterious mutations of HNRNP genes result in shared neurodevelopmental disorders.Genome Med. 2021 Apr 19;13(1):63. doi: 10.1186/s13073-021-00870-6. Genome Med. 2021. PMID: 33874999 Free PMC article.
-
CAGI6 ID panel challenge: assessment of phenotype and variant predictions in 415 children with neurodevelopmental disorders (NDDs).Hum Genet. 2025 Mar;144(2-3):227-242. doi: 10.1007/s00439-024-02722-w. Epub 2025 Jan 9. Hum Genet. 2025. PMID: 39786577 Free PMC article.
-
Design and Use of Semantic Resources: Findings from the Section on Knowledge Representation and Management of the 2020 International Medical Informatics Association Yearbook.Yearb Med Inform. 2020 Aug;29(1):163-168. doi: 10.1055/s-0040-1702010. Epub 2020 Aug 21. Yearb Med Inform. 2020. PMID: 32823311 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

