Cryptosporidium parvum and Cryptosporidium hominis subtypes in crab-eating macaques
- PMID: 31307508
- PMCID: PMC6631616
- DOI: 10.1186/s13071-019-3604-7
Cryptosporidium parvum and Cryptosporidium hominis subtypes in crab-eating macaques
Abstract
Background: Non-human primates are often infected with human-pathogenic Cryptosporidium hominis subtypes, but rarely with Cryptosporidium parvum. In this study, 1452 fecal specimens were collected from farmed crab-eating macaques (Macaca fascicularis) in Hainan, China during the period April 2016 to January 2018. These specimens were analyzed for Cryptosporidium species and subtypes by using PCR and sequence analysis of the 18S rRNA and 60 kDa glycoprotein (gp60) genes, respectively.
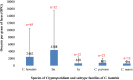
Results: Altogether, Cryptosporidium was detected using 18S rRNA-based PCR in 132 (9.1%) sampled animals, with significantly higher prevalence in females (12.5% or 75/599 versus 6.1% or 43/706), younger animals (10.7% or 118/1102 in monkeys 1-3-years-old versus 4.0% or 14/350 in those over 3-years-old) and animals with diarrhea (12.6% or 46/365 versus 7.9% or 86/1087). Four Cryptosporidium species were identified, namely C. hominis, C. parvum, Cryptosporidium muris and Cryptosporidium ubiquitum in 86, 30, 15 and 1 animal, respectively. The identified C. parvum, C. hominis and C. ubiquitum were further subtyped by using gp60 PCR. Among them, C. parvum belonged to subtypes in two known subtype families, namely IIoA14G1 (in 18 animals) and IIdA19G1 (in 2 animals). In contrast, C. hominis mostly belonged to two new subtype families Im and In, which are genetically related to Ia and Id, respectively. The C. hominis subtypes identified included ImA18 (in 38 animals), InA14 (in six animals), InA26 (in six animals), InA17 (in one animal) and IiA17 (in three animals). The C. ubiquitum isolates belonged to subtype family XIId. By subtype, ImA18 and IIoA14G1 were detected in animals with diarrhea whereas the remaining ones were mostly found in asymptomatic animals. Compared with C. parvum and C. muris, higher oocyst shedding intensity was observed in animals infected with C. hominis, especially those infected with the Im subtype family.
Conclusions: Data from the study suggest that crab-eating macaques are infected with diverse C. parvum and C. hominis subtypes. The C. parvum IIo subtype family previously seen in rodents in China has apparently expanded its host range.
Keywords: Crab-eating macaques; Cryptosporidium; Cryptosporidium hominis; Cryptosporidium parvum; Subtypes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Common occurrence of divergent Cryptosporidium species and Cryptosporidium parvum subtypes in farmed bamboo rats (Rhizomys sinensis).Parasit Vectors. 2020 Mar 24;13(1):149. doi: 10.1186/s13071-020-04021-5. Parasit Vectors. 2020. PMID: 32204732 Free PMC article.
-
Molecular prevalence and subtyping of Cryptosporidium hominis among captive long-tailed macaques (Macaca fascicularis) and rhesus macaques (Macaca mulatta) from Hainan Island, southern China.Parasit Vectors. 2019 Apr 30;12(1):192. doi: 10.1186/s13071-019-3449-0. Parasit Vectors. 2019. PMID: 31039801 Free PMC article.
-
First molecular characterization of Cryptosporidium in Yemen.Parasitology. 2013 May;140(6):729-34. doi: 10.1017/S0031182012001953. Epub 2013 Feb 1. Parasitology. 2013. PMID: 23369243
-
A review of recent Cryptosporidium hominis and Cryptosporidium parvum gp60 subtypes.Curr Res Parasitol Vector Borne Dis. 2025 Jul 6;8:100292. doi: 10.1016/j.crpvbd.2025.100292. eCollection 2025. Curr Res Parasitol Vector Borne Dis. 2025. PMID: 40734660 Free PMC article. Review.
-
Genomics and population biology of Cryptosporidium species.Parasite Immunol. 2012 Feb-Mar;34(2-3):61-71. doi: 10.1111/j.1365-3024.2011.01301.x. Parasite Immunol. 2012. PMID: 21595702 Free PMC article. Review.
Cited by
-
High zoonotic potential of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in wild nonhuman primates from Yunnan Province, China.Parasit Vectors. 2022 Mar 12;15(1):85. doi: 10.1186/s13071-022-05217-7. Parasit Vectors. 2022. PMID: 35279196 Free PMC article.
-
An annotated checklist of the eukaryotic parasites of humans, exclusive of fungi and algae.Zookeys. 2021 Nov 9;1069:1-313. doi: 10.3897/zookeys.1069.67403. eCollection 2021. Zookeys. 2021. PMID: 34819766 Free PMC article.
-
Molecular characterization of the waterborne pathogens Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. in wastewater and sewage in Guangzhou, China.Parasit Vectors. 2021 Jan 20;14(1):66. doi: 10.1186/s13071-020-04566-5. Parasit Vectors. 2021. PMID: 33472683 Free PMC article.
-
Cryptosporidium Species and C. parvum Subtypes in Farmed Bamboo Rats.Pathogens. 2020 Dec 2;9(12):1018. doi: 10.3390/pathogens9121018. Pathogens. 2020. PMID: 33276616 Free PMC article.
-
Exploratory Algorithm of a Multi-epitope-based Subunit Vaccine Candidate Against Cryptosporidium hominis: Reverse Vaccinology-Based Immunoinformatic Approach.Int J Pept Res Ther. 2022;28(5):134. doi: 10.1007/s10989-022-10438-6. Epub 2022 Jul 26. Int J Pept Res Ther. 2022. PMID: 35911179 Free PMC article.
References
-
- Li J, Dong H, Wang R, Yu F, Wu Y, Chang Y, et al. An investigation of parasitic infections and review of molecular characterization of the intestinal protozoa in nonhuman primates in China from 2009 to 2015. Int J Parasitol Parasites Wildl. 2017;6:8–15. doi: 10.1016/j.ijppaw.2016.12.003. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

