Impact of Antibiotic Gut Exposure on the Temporal Changes in Microbiome Diversity
- PMID: 31307985
- PMCID: PMC6761552
- DOI: 10.1128/AAC.00820-19
Impact of Antibiotic Gut Exposure on the Temporal Changes in Microbiome Diversity
Abstract
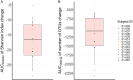
Although the global deleterious impact of antibiotics on the intestinal microbiota is well known, temporal changes in microbial diversity during and after an antibiotic treatment are still poorly characterized. We used plasma and fecal samples collected frequently during treatment and up to one month after from 22 healthy volunteers assigned to a 5-day treatment by moxifloxacin (n = 14) or no intervention (n = 8). Moxifloxacin concentrations were measured in both plasma and feces, and bacterial diversity was determined in feces by 16S rRNA gene profiling and quantified using the Shannon index and number of operational taxonomic units (OTUs). Nonlinear mixed effect models were used to relate drug pharmacokinetics and bacterial diversity over time. Moxifloxacin reduced bacterial diversity in a concentration-dependent manner, with a median maximal loss of 27.5% of the Shannon index (minimum [min], 17.5; maximum [max], 27.7) and 47.4% of the number of OTUs (min, 30.4; max, 48.3). As a consequence of both the long fecal half-life of moxifloxacin and the susceptibility of the gut microbiota to moxifloxacin, bacterial diversity indices did not return to their pretreatment levels until days 16 and 21, respectively. Finally, the model characterized the effect of moxifloxacin on bacterial diversity biomarkers and provides a novel framework for analyzing antibiotic effects on the intestinal microbiome.
Keywords: diversity; dysbiosis; intestinal microbiome; metagenomics; moxifloxacin; nonlinear mixed-effect modelling.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Rosen CE, Palm NW. 2017. Functional classification of the gut microbiota: the key to cracking the microbiota composition code: functional classifications of the gut microbiota reveal previously hidden contributions of indigenous gut bacteria to human health and disease. Bioessays 39:1700032. doi:10.1002/bies.201700032. - DOI - PubMed
-
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi:10.1056/NEJMoa1205037. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

