Identifying New Substrates and Functions for an Old Enzyme: Calcineurin
- PMID: 31308145
- PMCID: PMC7050593
- DOI: 10.1101/cshperspect.a035436
Identifying New Substrates and Functions for an Old Enzyme: Calcineurin
Abstract
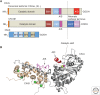
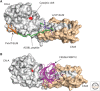
Biological processes are dynamically regulated by signaling networks composed of protein kinases and phosphatases. Calcineurin, or PP3, is a conserved phosphoserine/phosphothreonine-specific protein phosphatase and member of the PPP family of phosphatases. Calcineurin is unique, however, in its activation by Ca2+ and calmodulin. This ubiquitously expressed phosphatase controls Ca2+-dependent processes in all human tissues, but is best known for driving the adaptive immune response by dephosphorylating the nuclear factor of the activated T-cells (NFAT) family of transcription factors. Therefore, calcineurin inhibitors, FK506 (tacrolimus), and cyclosporin A serve as immunosuppressants. We describe some of the adverse effects associated with calcineurin inhibitors that result from inhibition of calcineurin in nonimmune tissues, illustrating the many functions of this enzyme that have yet to be elucidated. In fact, calcineurin has essential roles beyond the immune system, from yeast to humans, but since its discovery more than 30 years ago, only a small number of direct calcineurin substrates have been shown (∼75 proteins). This is because of limitations in current methods for identification of phosphatase substrates. Here we discuss recent insights into mechanisms of calcineurin activation and substrate recognition that have been critical in the development of novel approaches for identifying its targets systematically. Rather than comprehensively reviewing known functions of calcineurin, we highlight new approaches to substrate identification for this critical regulator that may reveal molecular mechanisms underlying toxicities caused by calcineurin inhibitor-based immunosuppression.
Copyright © 2020 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
-
- Alvaro CG, O'Donnell AF, Prosser DC, Augustine AA, Goldman A, Brodsky JL, Cyert MS, Wendland B, Thorner J. 2014. Specific α-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol Cell Biol 34: 2660–2681. 10.1128/MCB.00230-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
