The RNA exosome nuclease complex regulates human embryonic stem cell differentiation
- PMID: 31308215
- PMCID: PMC6683745
- DOI: 10.1083/jcb.201811148
The RNA exosome nuclease complex regulates human embryonic stem cell differentiation
Abstract
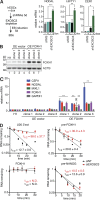
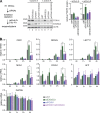
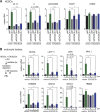
A defining feature of embryonic stem cells (ESCs) is the ability to differentiate into all three germ layers. Pluripotency is maintained in part by a unique transcription network that maintains expression of pluripotency-specific transcription factors and represses developmental genes. While the mechanisms that establish this transcription network are well studied, little is known of the posttranscriptional surveillance pathways that degrade differentiation-related RNAs. We report that the surveillance pathway mediated by the RNA exosome nuclease complex represses ESC differentiation. Depletion of the exosome expedites differentiation of human ESCs into all three germ layers. LINE-1 retrotransposons and specific miRNAs, lncRNAs, and mRNAs that encode developmental regulators or affect their expression are all bound by the exosome and increase in level upon exosome depletion. The exosome restrains differentiation in part by degrading transcripts encoding FOXH1, a transcription factor crucial for mesendoderm formation. Our studies establish the exosome as a regulator of human ESC differentiation and reveal the importance of RNA decay in maintaining pluripotency.
© 2019 Belair et al.
Figures
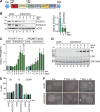

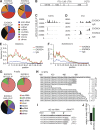
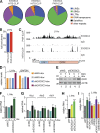

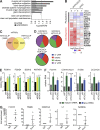
Comment in
-
A precision RNA degradation machinery shapes stem cell development.J Cell Biol. 2019 Aug 5;218(8):2437-2438. doi: 10.1083/jcb.201906115. Epub 2019 Jul 19. J Cell Biol. 2019. PMID: 31324648 Free PMC article.
Similar articles
-
Tuning FOXD3 expression dose-dependently balances human embryonic stem cells between pluripotency and meso-endoderm fates.Biochim Biophys Acta Mol Cell Res. 2019 Dec;1866(12):118531. doi: 10.1016/j.bbamcr.2019.118531. Epub 2019 Aug 12. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 31415841
-
Inhibition of MTA2 and MTA3 induces mesendoderm specification of human embryonic stem cells.Biochem Biophys Res Commun. 2021 May 7;552:142-149. doi: 10.1016/j.bbrc.2021.03.030. Epub 2021 Mar 19. Biochem Biophys Res Commun. 2021. PMID: 33744762
-
miR-373 is regulated by TGFβ signaling and promotes mesendoderm differentiation in human Embryonic Stem Cells.Dev Biol. 2014 Jul 1;391(1):81-8. doi: 10.1016/j.ydbio.2014.03.020. Epub 2014 Apr 4. Dev Biol. 2014. PMID: 24709321 Free PMC article.
-
Pluripotency Crossroads: Junction of Transcription Factors, Epigenetic Mechanisms, MicroRNAs, and Long Non-coding RNAs.Curr Stem Cell Res Ther. 2017;12(4):300-311. doi: 10.2174/1574888X12666170216155850. Curr Stem Cell Res Ther. 2017. PMID: 25986623 Review.
-
Post-transcriptional control of cellular differentiation by the RNA exosome complex.Nucleic Acids Res. 2020 Dec 2;48(21):11913-11928. doi: 10.1093/nar/gkaa883. Nucleic Acids Res. 2020. PMID: 33119769 Free PMC article. Review.
Cited by
-
Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery.Nat Rev Mol Cell Biol. 2020 Mar;21(3):123-136. doi: 10.1038/s41580-019-0209-0. Epub 2020 Feb 4. Nat Rev Mol Cell Biol. 2020. PMID: 32020081 Free PMC article. Review.
-
RNA-binding protein Mub1 and the nuclear RNA exosome act to fine-tune environmental stress response.Life Sci Alliance. 2021 Nov 30;5(2):e202101111. doi: 10.26508/lsa.202101111. Print 2022 Feb. Life Sci Alliance. 2021. PMID: 34848435 Free PMC article.
-
Control of RNA degradation in cell fate decision.Front Cell Dev Biol. 2023 Mar 21;11:1164546. doi: 10.3389/fcell.2023.1164546. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37025171 Free PMC article. Review.
-
RNA degradation eliminates developmental transcripts during murine embryonic stem cell differentiation via CAPRIN1-XRN2.Dev Cell. 2022 Dec 19;57(24):2731-2744.e5. doi: 10.1016/j.devcel.2022.11.014. Epub 2022 Dec 9. Dev Cell. 2022. PMID: 36495875 Free PMC article.
-
A precision RNA degradation machinery shapes stem cell development.J Cell Biol. 2019 Aug 5;218(8):2437-2438. doi: 10.1083/jcb.201906115. Epub 2019 Jul 19. J Cell Biol. 2019. PMID: 31324648 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

