Nitrogen sourcing during viral infection of marine cyanobacteria
- PMID: 31308237
- PMCID: PMC6681717
- DOI: 10.1073/pnas.1901856116
Nitrogen sourcing during viral infection of marine cyanobacteria
Abstract
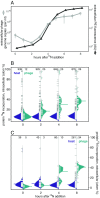
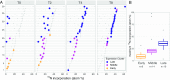
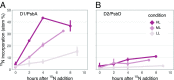
The building blocks of a virus derived from de novo biosynthesis during infection and/or catabolism of preexisting host cell biomass, and the relative contribution of these 2 sources has important consequences for understanding viral biogeochemistry. We determined the uptake of extracellular nitrogen (N) and its biosynthetic incorporation into both virus and host proteins using an isotope-labeling proteomics approach in a model marine cyanobacterium Synechococcus WH8102 infected by a lytic cyanophage S-SM1. By supplying dissolved N as 15N postinfection, we found that proteins in progeny phage particles were composed of up to 41% extracellularly derived N, while proteins of the infected host cell showed almost no isotope incorporation, demonstrating that de novo amino acid synthesis continues during infection and contributes specifically and substantially to phage replication. The source of N for phage protein synthesis shifted over the course of infection from mostly host derived in the early stages to more medium derived later on. We show that the photosystem II reaction center proteins D1 and D2, which are auxiliary metabolic genes (AMGs) in the S-SM1 genome, are made de novo during infection in an apparently light-dependent manner. We also identified a small set of host proteins that continue to be produced during infection; the majority are homologs of AMGs in S-SM1 or other viruses, suggesting selective continuation of host protein production during infection. The continued acquisition of nutrients by the infected cell and their utilization for phage replication are significant for both evolution and biogeochemical impact of viruses.
Keywords: bacteriophage; biogeochemistry; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Cyanophage infection and photoinhibition in marine cyanobacteria.Res Microbiol. 2004 Nov;155(9):720-5. doi: 10.1016/j.resmic.2004.06.002. Res Microbiol. 2004. PMID: 15501648 Review.
-
Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts.PLoS Biol. 2006 Jul;4(8):e234. doi: 10.1371/journal.pbio.0040234. PLoS Biol. 2006. PMID: 16802857 Free PMC article.
-
Shedding new light on viral photosynthesis.Photosynth Res. 2015 Oct;126(1):71-97. doi: 10.1007/s11120-014-0057-x. Epub 2014 Nov 9. Photosynth Res. 2015. PMID: 25381655 Review.
-
Characterization of mutants expressing thermostable D1 and D2 polypeptides of photosystem II in the cyanobacterium Synechococcus elongatus PCC 7942.J Biosci Bioeng. 2018 Oct;126(4):417-424. doi: 10.1016/j.jbiosc.2018.04.015. Epub 2018 Jun 8. J Biosci Bioeng. 2018. PMID: 29891421
-
Two Synechococcus genes, Two Different Effects on Cyanophage Infection.Viruses. 2017 Jun 2;9(6):136. doi: 10.3390/v9060136. Viruses. 2017. PMID: 28574452 Free PMC article.
Cited by
-
Viruses affect picocyanobacterial abundance and biogeography in the North Pacific Ocean.Nat Microbiol. 2022 Apr;7(4):570-580. doi: 10.1038/s41564-022-01088-x. Epub 2022 Apr 1. Nat Microbiol. 2022. PMID: 35365792 Free PMC article.
-
A novel roseosiphovirus infecting dinoroseobacter shibae DFL12T represents a new genus.BMC Genomics. 2025 Feb 8;26(1):121. doi: 10.1186/s12864-025-11274-w. BMC Genomics. 2025. PMID: 39923004 Free PMC article.
-
Deciphering the Virus Signal Within the Marine Dissolved Organic Matter Pool.Front Microbiol. 2022 May 27;13:863686. doi: 10.3389/fmicb.2022.863686. eCollection 2022. Front Microbiol. 2022. PMID: 35694303 Free PMC article.
-
Deciphering Active Prophages from Metagenomes.mSystems. 2022 Apr 26;7(2):e0008422. doi: 10.1128/msystems.00084-22. Epub 2022 Mar 24. mSystems. 2022. PMID: 35323045 Free PMC article.
-
Identification of Novel Viruses and Their Microbial Hosts from Soils with Long-Term Nitrogen Fertilization and Cover Cropping Management.mSystems. 2022 Dec 20;7(6):e0057122. doi: 10.1128/msystems.00571-22. Epub 2022 Nov 29. mSystems. 2022. PMID: 36445691 Free PMC article.
References
-
- Danovaro R., et al. , Marine viruses and global climate change. FEMS Microbiol. Rev. 35, 993–1034 (2011). - PubMed
-
- Weitz J. S., Quantitative Viral Ecology: Dynamics of Viruses and Their Microbial Hosts (Princeton University Press, 2016).
-
- Mateus M. D., Bridging the gap between knowing and modeling viruses in marine systems—An upcoming frontier. Front. Mar. Sci. 3, 284 (2017).
-
- Motegi C., Kaiser K., Benner R., Weinbauer M. G., Effect of P-limitation on prokaryotic and viral production in surface waters of the Northwestern Mediterranean Sea. J. Plankton Res. 37, 16–20 (2015).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

