Detection of cell-type-specific risk-CpG sites in epigenome-wide association studies
- PMID: 31308366
- PMCID: PMC6629651
- DOI: 10.1038/s41467-019-10864-z
Detection of cell-type-specific risk-CpG sites in epigenome-wide association studies
Abstract
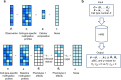
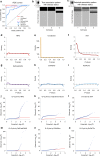
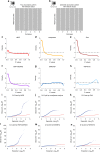
In epigenome-wide association studies, the measured signals for each sample are a mixture of methylation profiles from different cell types. Current approaches to the association detection claim whether a cytosine-phosphate-guanine (CpG) site is associated with the phenotype or not at aggregate level and can suffer from low statistical power. Here, we propose a statistical method, HIgh REsolution (HIRE), which not only improves the power of association detection at aggregate level as compared to the existing methods but also enables the detection of risk-CpG sites for individual cell types.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
DNA methylation in childhood asthma: an epigenome-wide meta-analysis.Lancet Respir Med. 2018 May;6(5):379-388. doi: 10.1016/S2213-2600(18)30052-3. Epub 2018 Feb 26. Lancet Respir Med. 2018. PMID: 29496485
-
Epigenome-wide association study of DNA methylation and adult asthma in the Agricultural Lung Health Study.Eur Respir J. 2020 Sep 3;56(3):2000217. doi: 10.1183/13993003.00217-2020. Print 2020 Sep. Eur Respir J. 2020. PMID: 32381493 Free PMC article.
-
Correcting for Sample Heterogeneity in Methylome-Wide Association Studies.Methods Mol Biol. 2017;1589:107-114. doi: 10.1007/7651_2015_266. Methods Mol Biol. 2017. PMID: 26246354
-
Epigenome-wide association studies in asthma: A systematic review.Clin Exp Allergy. 2019 Jul;49(7):953-968. doi: 10.1111/cea.13403. Epub 2019 May 14. Clin Exp Allergy. 2019. PMID: 31009112
-
Recommendations for the design and analysis of epigenome-wide association studies.Nat Methods. 2013 Oct;10(10):949-55. doi: 10.1038/nmeth.2632. Nat Methods. 2013. PMID: 24076989 Review.
Cited by
-
CeDAR: incorporating cell type hierarchy improves cell type-specific differential analyses in bulk omics data.Genome Biol. 2023 Feb 28;24(1):37. doi: 10.1186/s13059-023-02857-5. Genome Biol. 2023. PMID: 36855165 Free PMC article.
-
A comprehensive assessment of cell type-specific differential expression methods in bulk data.Brief Bioinform. 2023 Jan 19;24(1):bbac516. doi: 10.1093/bib/bbac516. Brief Bioinform. 2023. PMID: 36472568 Free PMC article.
-
iProMix: A mixture model for studying the function of ACE2 based on bulk proteogenomic data.J Am Stat Assoc. 2023;118(541):43-55. doi: 10.1080/01621459.2022.2110876. Epub 2022 Oct 5. J Am Stat Assoc. 2023. PMID: 37409267 Free PMC article.
-
A pan-tissue DNA methylation atlas enables in silico decomposition of human tissue methylomes at cell-type resolution.Nat Methods. 2022 Mar;19(3):296-306. doi: 10.1038/s41592-022-01412-7. Epub 2022 Mar 11. Nat Methods. 2022. PMID: 35277705 Free PMC article.
-
Cell-type-specific resolution epigenetics without the need for cell sorting or single-cell biology.Nat Commun. 2019 Jul 31;10(1):3417. doi: 10.1038/s41467-019-11052-9. Nat Commun. 2019. PMID: 31366909 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

