HLA class II-Restricted CD8+ T cells in HIV-1 Virus Controllers
- PMID: 31308388
- PMCID: PMC6629643
- DOI: 10.1038/s41598-019-46462-8
HLA class II-Restricted CD8+ T cells in HIV-1 Virus Controllers
Abstract
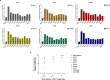
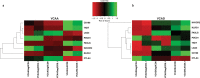
A paradigm shifting study demonstrated that induction of MHC class E and II-restricted CD8+ T cells was associated with the clearance of SIV infection in rhesus macaques. Another recent study highlighted the presence of HIV-1-specific class II-restricted CD8+ T cells in HIV-1 patients who naturally control infection (virus controllers; VCs). However, questions regarding class II-restricted CD8+ T cells ontogeny, distribution across different HIV-1 disease states and their role in viral control remain unclear. In this study, we investigated the distribution and anti-viral properties of HLA-DRB1*0701 and DQB1*0501 class II-restricted CD8+ T cells in different HIV-1 patient cohorts; and whether class II-restricted CD8+ T cells represent a unique T cell subset. We show that memory class II-restricted CD8+ T cell responses were more often detectable in VCs than in chronically infected patients, but not in healthy seronegative donors. We also demonstrate that VC CD8+ T cells inhibit virus replication in both a class I- and class II-dependent manner, and that in two VC patients the class II-restricted CD8+ T cells with an anti-viral gene signature expressed both CD4+ and CD8+ T cell lineage-specific genes. These data demonstrated that anti-viral memory class II-restricted CD8+ T cells with hybrid CD4+ and CD8+ features are present during natural HIV-1 infection.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
CD8+ T Cell Breadth and Ex Vivo Virus Inhibition Capacity Distinguish between Viremic Controllers with and without Protective HLA Class I Alleles.J Virol. 2016 Jul 11;90(15):6818-6831. doi: 10.1128/JVI.00276-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194762 Free PMC article.
-
HIV controllers with HLA-DRB1*13 and HLA-DQB1*06 alleles have strong, polyfunctional mucosal CD4+ T-cell responses.J Virol. 2010 Nov;84(21):11020-9. doi: 10.1128/JVI.00980-10. Epub 2010 Aug 18. J Virol. 2010. PMID: 20719952 Free PMC article.
-
Acute-Phase CD4+ T Cell Responses Targeting Invariant Viral Regions Are Associated with Control of Live Attenuated Simian Immunodeficiency Virus.J Virol. 2018 Oct 12;92(21):e00830-18. doi: 10.1128/JVI.00830-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111562 Free PMC article.
-
Mucosal immunity in HIV controllers: the right place at the right time.Curr Opin HIV AIDS. 2011 May;6(3):202-7. doi: 10.1097/COH.0b013e3283453e2b. Curr Opin HIV AIDS. 2011. PMID: 21399497 Free PMC article. Review.
-
Immunologic Control of HIV-1: What Have We Learned and Can We Induce It?Curr HIV/AIDS Rep. 2021 Jun;18(3):211-220. doi: 10.1007/s11904-021-00545-2. Epub 2021 Mar 11. Curr HIV/AIDS Rep. 2021. PMID: 33709324 Review.
Cited by
-
A class-mismatched TCR bypasses MHC restriction via an unorthodox but fully functional binding geometry.Nat Commun. 2022 Nov 23;13(1):7189. doi: 10.1038/s41467-022-34896-0. Nat Commun. 2022. PMID: 36424374 Free PMC article.
-
The Evolving T Cell Receptor Recognition Code: The Rules Are More Like Guidelines.Immunol Rev. 2025 Jan;329(1):e13439. doi: 10.1111/imr.13439. Immunol Rev. 2025. PMID: 39804137 Review.
-
Polyclonal Broadly Neutralizing Antibody Activity Characterized by CD4 Binding Site and V3-Glycan Antibodies in a Subset of HIV-1 Virus Controllers.Front Immunol. 2021 Dec 23;12:670561. doi: 10.3389/fimmu.2021.670561. eCollection 2021. Front Immunol. 2021. PMID: 35003053 Free PMC article.
-
HIV-1 adapts to HLA class II-associated selection pressure exerted by CD4+ and CD8+ T cells.Sci Adv. 2025 Feb 14;11(7):eadr4238. doi: 10.1126/sciadv.adr4238. Epub 2025 Feb 14. Sci Adv. 2025. PMID: 39951541 Free PMC article.
-
Human CD4 T Cells From Thymus and Cord Blood Are Convertible Into CD8 T Cells by IL-4.Front Immunol. 2022 Feb 11;13:834033. doi: 10.3389/fimmu.2022.834033. eCollection 2022. Front Immunol. 2022. PMID: 35222424 Free PMC article.
References
-
- UNAIDS. FACT SHEET NOVEMBER 2016, http://www.unaids.org/en/resources/fact-sheet (2016).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

