REV-ERBα and REV-ERBβ function as key factors regulating Mammalian Circadian Output
- PMID: 31308426
- PMCID: PMC6629614
- DOI: 10.1038/s41598-019-46656-0
REV-ERBα and REV-ERBβ function as key factors regulating Mammalian Circadian Output
Abstract
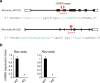
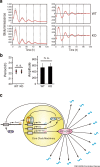
The circadian clock regulates behavioural and physiological processes in a 24-h cycle. The nuclear receptors REV-ERBα and REV-ERBβ are involved in the cell-autonomous circadian transcriptional/translational feedback loops as transcriptional repressors. A number of studies have also demonstrated a pivotal role of REV-ERBs in regulation of metabolic, neuronal, and inflammatory functions including bile acid metabolism, lipid metabolism, and production of inflammatory cytokines. Given the multifunctional role of REV-ERBs, it is important to elucidate the mechanism through which REV-ERBs exert their functions. To this end, we established a Rev-erbα/Rev-erbβ double-knockout mouse embryonic stem (ES) cell model and analyzed the circadian clock and clock-controlled output gene expressions. A comprehensive mRNA-seq analysis revealed that the double knockout of both Rev-erbα and Rev-erbβ does not abrogate expression rhythms of E-box-regulated core clock genes but drastically changes a diverse set of other rhythmically-expressed output genes. Of note, REV-ERBα/β deficiency does not compromise circadian expression rhythms of PER2, while REV-ERB target genes, Bmal1 and Npas2, are significantly upregulated. This study highlight the relevance of REV-ERBs as pivotal output mediators of the mammalian circadian clock.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

