Predicting Antigenicity of Influenza A Viruses Using biophysical ideas
- PMID: 31308446
- PMCID: PMC6629677
- DOI: 10.1038/s41598-019-46740-5
Predicting Antigenicity of Influenza A Viruses Using biophysical ideas
Abstract
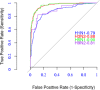
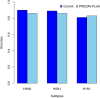
Antigenic variations of influenza A viruses are induced by genomic mutation in their trans-membrane protein HA1, eliciting viral escape from neutralization by antibodies generated in prior infections or vaccinations. Prediction of antigenic relationships among influenza viruses is useful for designing (or updating the existing) influenza vaccines, provides important insights into the evolutionary mechanisms underpinning viral antigenic variations, and helps to understand viral epidemiology. In this study, we present a simple and physically interpretable model that can predict antigenic relationships among influenza A viruses, based on biophysical ideas, using both genomic amino acid sequences and experimental antigenic data. We demonstrate the applicability of the model using a benchmark dataset of four subtypes of influenza A (H1N1, H3N2, H5N1, and H9N2) viruses and report on its performance profiles. Additionally, analysis of the model's parameters confirms several observations that are consistent with the findings of other previous studies, for which we provide plausible explanations.
Conflict of interest statement
The authors declare no competing interests.
Figures

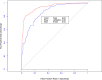
References
-
- Organization-(WHO), W. H. Influenza (Seasonal) Fact Sheets. WHO http://www.who.int/mediacentre/factsheets/fs211/en/ (2016).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

