Glycosylation of acyl carrier protein-bound polyketides during pactamycin biosynthesis
- PMID: 31308531
- PMCID: PMC6642016
- DOI: 10.1038/s41589-019-0314-6
Glycosylation of acyl carrier protein-bound polyketides during pactamycin biosynthesis
Abstract
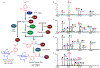
Glycosylation is a common modification reaction in natural product biosynthesis and has been known to be a post-assembly line tailoring process in glycosylated polyketide biosynthesis. Here, we show that in pactamycin biosynthesis, glycosylation can take place on an acyl carrier protein (ACP)-bound polyketide intermediate. Using in vivo gene inactivation, chemical complementation and in vitro pathway reconstitution, we demonstrate that the 3-aminoacetophenone moiety of pactamycin is derived from 3-aminobenzoic acid by a set of discrete polyketide synthase proteins via a 3-(3-aminophenyl)3-oxopropionyl-ACP intermediate. This ACP-bound intermediate is then glycosylated by an N-glycosyltransferase, PtmJ, providing a sugar precursor for the formation of the aminocyclopentitol core structure of pactamycin. This is the first example of glycosylation of a small molecule while tethered to a carrier protein. Additionally, we demonstrate that PtmO is a hydrolase that is responsible for the release of the ACP-bound product to a free β-ketoacid that subsequently undergoes decarboxylation.
Conflict of interest statement
Competing Interests
The authors declare no competing financial interest.
Figures




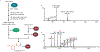
References
-
- Mao Y, Varoglu M & Sherman DH Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem Biol 6, 251–263 (1999). - PubMed
-
- Kudo F, Kasama Y, Hirayama T & Eguchi T Cloning of the pactamycin biosynthetic gene cluster and characterization of a crucial glycosyltransferase prior to a unique cyclopentane ring formation. J Antibiot 60, 492–503 (2007). - PubMed
-
- Ito T et al. Deciphering pactamycin biosynthesis and engineered production of new pactamycin analogues. ChemBioChem 10, 2253–2265 (2009). - PubMed
-
- Rinehart KL Jr., Weller DD & Pearce CJ Recent biosynthetic studies on antibiotics. J. Nat. Prod 43, 1–20 (1980).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

