Molecular variability of Streptococcus uberis isolates from intramammary infections in Canadian dairy farms from the Maritime region
- PMID: 31308588
- PMCID: PMC6587884
Molecular variability of Streptococcus uberis isolates from intramammary infections in Canadian dairy farms from the Maritime region
Abstract
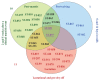
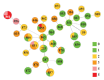
The primary objective of this study was to explore the variability of Streptococcus uberis (S. uberis) isolates by extracting multilocus sequence typing (MLST) data from whole-genome sequencing. The secondary objective was to determine the distribution of the phenotypic antimicrobial resistance (AMR) and the associated AMR genes as well as the virulence gene profiles among sequence types (STs). Sixty-two isolates were recovered from 16 herds in 3 Canadian Maritime Provinces: New Brunswick (14.5%), Nova Scotia (48.3%), and Prince Edward Island (37.1%). Of these, 9, 30, and 23 were recovered from post-calving, lactational samples, and post-mastitis samples, respectively. These 62 S. uberis isolates belonged to 34 STs; 11 isolates were typed to 9 known STs and 51 isolates were classified as one of 25 new STs. Thirteen isolates were part of major clonal complexes (CCs). Post-mastitis isolates contained 10 unique STs, lactational isolates contained 11 unique STs, and post-calving isolates had 3 STs. Each farm had only 1 isolate that was a unique ST except for STs 233, 851, 855, 857, 864, and 866, which were found in multiple cows per herd on more than one farm. ST851 and ST857 were found in each of the 3 sample types, with ST857 found in cows from all 3 Maritime provinces. These results indicate that S. uberis is a diverse non-clonal pathogen with specific STs residing in clonal clusters, carrying multiple AMR genes and virulence, with a diverse phenotypic AMR.
L’objectif premier de la présente étude était d’explorer la variabilité d’isolats de Streptococcus uberis en extrayant les données de typage génomique multilocus (MLST) du séquençage du génome entier. L’objectif secondaire était de déterminer la distribution des phénotypes de résistance antimicrobienne (AMR) et les gènes d’AMR associés ainsi que les profils des gènes de virulence parmi les types de séquence (STs). Soixante-deux isolats ont été obtenus de 16 troupeaux dans trois provinces maritimes canadiennes : le Nouveau-Brunswick (14,5 %), la Nouvelle-Écosse (48,3 %), et l’Ile-du Prince-Édouard (37,1 %). Parmi ces isolats, 9, 30, et 23 ont été obtenus d’échantillons post-vêlage, en lactation, et post-mammite, respectivement. Ces 62 échantillons appartenaient à 34 STs; 11 isolats ont été typés comme appartenant à 9 STs connus et 51 isolats ont été classifiés dans un des 25 nouveaux STs. Treize isolats faisaient partis de complexes clonaux majeurs (CCs). Les isolats post-mammite contenaient 10 STs uniques, les isolats de la période de lactation contenaient 11 STs uniques, et ceux de la période post-vêlage avaient 3 STs. Chaque ferme n’avait seulement qu’un isolat qui avait un ST unique sauf pour les STs 233, 851, 855, 857, 864, et 866 qui ont été retrouvés chez plusieurs vaches par troupeau sur plus d’une ferme. Les ST851 et ST857 ont été trouvés dans chacun des trois types d’échantillons, avec ST857 retrouvé dans des vaches des trois provinces maritimes. Ces résultats indiquent que S. uberis est un agent pathogène diversifié non-clonal avec des STs spécifiques résidant dans des groupements clonaux, arborant de multiples gènes de résistance antimicrobienne et de virulence, avec des phénotypes d’AMR variés.(Traduit par Docteur Serge Messier).
Figures

Similar articles
-
Comparison of the population structure of Streptococcus uberis mastitis isolates from Austrian small-scale dairy farms and a Slovakian large-scale farm.J Dairy Sci. 2020 Feb;103(2):1820-1830. doi: 10.3168/jds.2019-16930. Epub 2019 Dec 16. J Dairy Sci. 2020. PMID: 31837784
-
Molecular characterization of Streptococcus agalactiae and Streptococcus uberis isolates from bovine milk.Trop Anim Health Prod. 2012 Dec;44(8):1981-92. doi: 10.1007/s11250-012-0167-4. Epub 2012 May 16. Trop Anim Health Prod. 2012. PMID: 22588571
-
Molecular Epidemiology of Streptococcus uberis Clinical Mastitis in Dairy Herds: Strain Heterogeneity and Transmission.J Clin Microbiol. 2016 Jan;54(1):68-74. doi: 10.1128/JCM.01583-15. Epub 2015 Oct 21. J Clin Microbiol. 2016. PMID: 26491180 Free PMC article.
-
Whole-Genome Sequence Analysis of Antimicrobial Resistance Genes in Streptococcus uberis and Streptococcus dysgalactiae Isolates from Canadian Dairy Herds.Front Vet Sci. 2017 May 22;4:63. doi: 10.3389/fvets.2017.00063. eCollection 2017. Front Vet Sci. 2017. PMID: 28589129 Free PMC article.
-
The exploitation of the genome in the search for determinants of virulence in Streptococcus uberis.Vet Immunol Immunopathol. 2004 Aug;100(3-4):145-9. doi: 10.1016/j.vetimm.2004.04.004. Vet Immunol Immunopathol. 2004. PMID: 15207452 Review.
Cited by
-
Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm.Antibiotics (Basel). 2021 May 28;10(6):644. doi: 10.3390/antibiotics10060644. Antibiotics (Basel). 2021. PMID: 34071296 Free PMC article.
-
Genomic surveillance reveals antibiotic resistance gene transmission via phage recombinases within sheep mastitis-associated Streptococcus uberis.BMC Vet Res. 2022 Jul 7;18(1):264. doi: 10.1186/s12917-022-03341-1. BMC Vet Res. 2022. PMID: 35799261 Free PMC article.
-
A core genome multi-locus sequence typing scheme for Streptococcus uberis: an evolution in typing a genetically diverse pathogen.Microb Genom. 2024 Mar;10(3):001225. doi: 10.1099/mgen.0.001225. Microb Genom. 2024. PMID: 38512314 Free PMC article.
-
Multilocus Sequence Genotype Heterogeneity in Streptococcus uberis Isolated from Bovine Mastitis in the Czech Republic.Animals (Basel). 2022 Sep 7;12(18):2327. doi: 10.3390/ani12182327. Animals (Basel). 2022. PMID: 36139187 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
