VariantQC: a visual quality control report for variant evaluation
- PMID: 31309221
- PMCID: PMC7963085
- DOI: 10.1093/bioinformatics/btz560
VariantQC: a visual quality control report for variant evaluation
Abstract
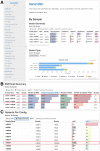
Summary: Large scale genomic studies produce millions of sequence variants, generating datasets far too massive for manual inspection. To ensure variant and genotype data are consistent and accurate, it is necessary to evaluate variants prior to downstream analysis using quality control (QC) reports. Variant call format (VCF) files are the standard format for representing variant data; however, generating summary statistics from these files is not always straightforward. While tools to summarize variant data exist, they generally produce simple text file tables, which still require additional processing and interpretation. VariantQC fills this gap as a user friendly, interactive visual QC report that generates and concisely summarizes statistics from VCF files. The report aggregates and summarizes variants by dataset, chromosome, sample and filter type. The VariantQC report is useful for high-level dataset summary, quality control and helps flag outliers. Furthermore, VariantQC operates on VCF files, so it can be easily integrated into many existing variant pipelines.
Availability and implementation: DISCVRSeq's VariantQC tool is freely available as a Java program, with the compiled JAR and source code available from https://github.com/BimberLab/DISCVRSeq/. Documentation and example reports are available at https://bimberlab.github.io/DISCVRSeq/.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
re-Searcher: GUI-based bioinformatics tool for simplified genomics data mining of VCF files.PeerJ. 2021 May 3;9:e11333. doi: 10.7717/peerj.11333. eCollection 2021. PeerJ. 2021. PMID: 33987016 Free PMC article.
-
Variant graph craft (VGC): a comprehensive tool for analyzing genetic variation and identifying disease-causing variants.BMC Bioinformatics. 2024 Sep 3;25(1):288. doi: 10.1186/s12859-024-05875-7. BMC Bioinformatics. 2024. PMID: 39227781 Free PMC article.
-
VCF-kit: assorted utilities for the variant call format.Bioinformatics. 2017 May 15;33(10):1581-1582. doi: 10.1093/bioinformatics/btx011. Bioinformatics. 2017. PMID: 28093408 Free PMC article.
-
Variant Tool Chest: an improved tool to analyze and manipulate variant call format (VCF) files.BMC Bioinformatics. 2014;15 Suppl 7(Suppl 7):S12. doi: 10.1186/1471-2105-15-S7-S12. Epub 2014 May 28. BMC Bioinformatics. 2014. PMID: 25080132 Free PMC article.
-
vcfView: An Extensible Data Visualization and Quality Assurance Platform for Integrated Somatic Variant Analysis.Cancer Inform. 2020 Nov 11;19:1176935120972377. doi: 10.1177/1176935120972377. eCollection 2020. Cancer Inform. 2020. PMID: 33239857 Free PMC article. Review.
Cited by
-
Extensive Phylogenomic Discordance and the Complex Evolutionary History of the Neotropical Cat Genus Leopardus.Mol Biol Evol. 2023 Dec 1;40(12):msad255. doi: 10.1093/molbev/msad255. Mol Biol Evol. 2023. PMID: 37987559 Free PMC article.
-
Whole Genome Sequencing Reveals Clade-Specific Genetic Variation in Blacklegged Ticks.Ecol Evol. 2025 Feb 11;15(2):e70987. doi: 10.1002/ece3.70987. eCollection 2025 Feb. Ecol Evol. 2025. PMID: 39944902 Free PMC article.
-
A quality control portal for sequencing data deposited at the European genome-phenome archive.Brief Bioinform. 2022 May 13;23(3):bbac136. doi: 10.1093/bib/bbac136. Brief Bioinform. 2022. PMID: 35438138 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

