Enzymes of early-diverging, zoosporic fungi
- PMID: 31309267
- PMCID: PMC6690862
- DOI: 10.1007/s00253-019-09983-w
Enzymes of early-diverging, zoosporic fungi
Abstract
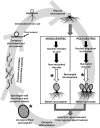
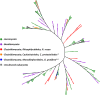
The secretome, the complement of extracellular proteins, is a reflection of the interaction of an organism with its host or substrate, thus a determining factor for the organism's fitness and competitiveness. Hence, the secretome impacts speciation and organismal evolution. The zoosporic Chytridiomycota, Blastocladiomycota, Neocallimastigomycota, and Cryptomycota represent the earliest diverging lineages of the Fungal Kingdom. The review describes the enzyme compositions of these zoosporic fungi, underscoring the enzymes involved in biomass degradation. The review connects the lifestyle and substrate affinities of the zoosporic fungi to the secretome composition by examining both classical phenotypic investigations and molecular/genomic-based studies. The carbohydrate-active enzyme profiles of 19 genome-sequenced species are summarized. Emphasis is given to recent advances in understanding the functional role of rumen fungi, the basis for the devastating chytridiomycosis, and the structure of fungal cellulosome. The approach taken by the review enables comparison of the secretome enzyme composition of anaerobic versus aerobic early-diverging fungi and comparison of enzyme portfolio of specialized parasites, pathogens, and saprotrophs. Early-diverging fungi digest most major types of biopolymers: cellulose, hemicellulose, pectin, chitin, and keratin. It is thus to be expected that early-diverging fungi in its entirety represents a rich and diverse pool of secreted, metabolic enzymes. The review presents the methods used for enzyme discovery, the diversity of enzymes found, the status and outlook for recombinant production, and the potential for applications. Comparative studies on the composition of secretome enzymes of early-diverging fungi would contribute to unraveling the basal lineages of fungi.
Keywords: Biomass-degrading enzymes; Biotech potential; Early-diverging fungi; Functional genomics; Secretome enzyme composition; Secretome evolution; Zoosporic aerobic and anaerobic fungi.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential.FEMS Microbiol Ecol. 2014 Oct;90(1):1-17. doi: 10.1111/1574-6941.12383. Epub 2014 Aug 11. FEMS Microbiol Ecol. 2014. PMID: 25046344 Review.
-
Cellulosome Localization Patterns Vary across Life Stages of Anaerobic Fungi.mBio. 2021 Jun 29;12(3):e0083221. doi: 10.1128/mBio.00832-21. Epub 2021 Jun 1. mBio. 2021. PMID: 34061594 Free PMC article.
-
The biotechnological potential of anaerobic fungi on fiber degradation and methane production.World J Microbiol Biotechnol. 2018 Oct 1;34(10):155. doi: 10.1007/s11274-018-2539-z. World J Microbiol Biotechnol. 2018. PMID: 30276481 Review.
-
Phylogenomic Analyses of Nucleotide-Sugar Biosynthetic and Interconverting Enzymes Illuminate Cell Wall Composition in Fungi.mBio. 2021 Apr 13;12(2):e03540-20. doi: 10.1128/mBio.03540-20. mBio. 2021. PMID: 33849982 Free PMC article.
-
Fungal bioconversion of lignocellulosic residues; opportunities & perspectives.Int J Biol Sci. 2009 Sep 4;5(6):578-95. doi: 10.7150/ijbs.5.578. Int J Biol Sci. 2009. PMID: 19774110 Free PMC article. Review.
Cited by
-
Mycoparasites, Gut Dwellers, and Saprotrophs: Phylogenomic Reconstructions and Comparative Analyses of Kickxellomycotina Fungi.Genome Biol Evol. 2023 Jan 4;15(1):evac185. doi: 10.1093/gbe/evac185. Genome Biol Evol. 2023. PMID: 36617272 Free PMC article.
-
Evolution of Fungal Carbohydrate-Active Enzyme Portfolios and Adaptation to Plant Cell-Wall Polymers.J Fungi (Basel). 2021 Mar 5;7(3):185. doi: 10.3390/jof7030185. J Fungi (Basel). 2021. PMID: 33807546 Free PMC article. Review.
-
Composition, Influencing Factors, and Effects on Host Nutrient Metabolism of Fungi in Gastrointestinal Tract of Monogastric Animals.Animals (Basel). 2025 Mar 1;15(5):710. doi: 10.3390/ani15050710. Animals (Basel). 2025. PMID: 40075993 Free PMC article. Review.
-
Mucilage protects the planktonic desmid Staurodesmus sp. against parasite attack by a chytrid fungus.J Plankton Res. 2022 Dec 27;45(1):3-14. doi: 10.1093/plankt/fbac071. eCollection 2023 Jan-Feb. J Plankton Res. 2022. PMID: 36751484 Free PMC article.
-
Succession of physiological stages hallmarks the transcriptomic response of the fungus Aspergillus niger to lignocellulose.Biotechnol Biofuels. 2020 Apr 13;13:69. doi: 10.1186/s13068-020-01702-2. eCollection 2020. Biotechnol Biofuels. 2020. PMID: 32313551 Free PMC article.
References
-
- Ahrendt SR, Quandt CA, Ciobanu D, Clum A, Salamov A, Andreopoulos B, Cheng JF, Woyke T, Pelin A, Henrissat B, Reynolds NK, Benny GL, Smith ME, James TY, Grigoriev IV. Leveraging single-cell genomics to expand the fungal tree of life. Nat Microbiol. 2018;3:1417–1428. doi: 10.1038/s41564-018-0261-0. - DOI - PMC - PubMed
-
- Benoit I, Coutinho PM, Schols HA, Gerlach JP, Henrissat B, de Vries RP. Degradation of different pectins by fungi: correlations and contrasts between pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC Genomics. 2012;13:321. doi: 10.1186/1471-2164-13-321. - DOI - PMC - PubMed
-
- Berger L, Roberts AA, Voyles J, Longcore JE, Murray KA, Skerratt History and recent progress on chytridiomycosis in amphibians. Fungal Ecol. 2016;19:89–99. doi: 10.1016/j.funeco.2015.09.007. - DOI
-
- Borneman S, Akin DE. The nature of anaerobic fungi and their polysaccharide degrading enzymes. Mycoscience. 1994;35:199–211. doi: 10.1007/BF02318501. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

