Templated chemistry for bioorganic synthesis and chemical biology
- PMID: 31309674
- PMCID: PMC6771651
- DOI: 10.1002/psc.3198
Templated chemistry for bioorganic synthesis and chemical biology
Abstract
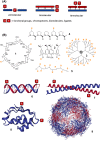
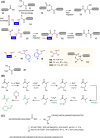
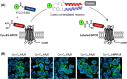
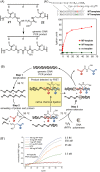
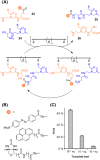
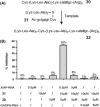
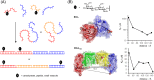
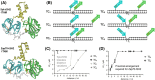
In light of the 2018 Max Bergmann Medal, this review discusses advancements on chemical biology-driven templated chemistry developed in the author's laboratories. The focused review introduces the template categories applied to orient functional units such as functional groups, chromophores, biomolecules, or ligands in space. Unimolecular templates applied in protein synthesis facilitate fragment coupling of unprotected peptides. Templating via bimolecular assemblies provides control over proximity relationships between functional units of two molecules. As an instructive example, the coiled coil peptide-templated labelling of receptor proteins on live cells will be shown. Termolecular assemblies provide the opportunity to put the proximity of functional units on two (bio)molecules under the control of a third party molecule. This allows the design of conditional bimolecular reactions. A notable example is DNA/RNA-triggered peptide synthesis. The last section shows how termolecular and multimolecular assemblies can be used to better characterize and understand multivalent protein-ligand interactions.
Keywords: Oligonucleotides; bioconjugation; carbohydrates; lectin; multivalency; peptide ligation; protein labeling; templated synthesis.
© 2019 The Authors Journal of Peptide Science published by European Peptide Society and John Wiley & Sons Ltd.
Figures



Similar articles
-
Templated native chemical ligation: peptide chemistry beyond protein synthesis.J Pept Sci. 2014 Feb;20(2):78-86. doi: 10.1002/psc.2602. Epub 2014 Jan 7. J Pept Sci. 2014. PMID: 24395765 Review.
-
A fluorogenic native chemical ligation for assessing the role of distance in peptide-templated peptide ligation.Bioorg Med Chem. 2017 Sep 15;25(18):5022-5030. doi: 10.1016/j.bmc.2017.08.007. Epub 2017 Aug 10. Bioorg Med Chem. 2017. PMID: 28823838
-
RNA-templated chemical synthesis of proapoptotic L- and d-peptides.Bioorg Med Chem. 2022 Jul 15;66:116786. doi: 10.1016/j.bmc.2022.116786. Epub 2022 May 14. Bioorg Med Chem. 2022. PMID: 35594647
-
PNA as a Biosupramolecular Tag for Programmable Assemblies and Reactions.Acc Chem Res. 2015 May 19;48(5):1319-31. doi: 10.1021/acs.accounts.5b00109. Acc Chem Res. 2015. PMID: 25947113
-
Templating Peptide Chemistry with Nucleic Acids: Toward Artificial Ribosomes, Cell-Specific Therapeutics, and Novel Protein-Mimetic Architectures.Biomacromolecules. 2024 Jul 8;25(7):3865-3876. doi: 10.1021/acs.biomac.4c00372. Epub 2024 Jun 11. Biomacromolecules. 2024. PMID: 38860980 Review.
Cited by
-
Assembly of Biologically Functional Structures by Nucleic Acid Templating: Implementation of a Strategy to Overcome Inhibition by Template Excess.Molecules. 2022 Oct 12;27(20):6831. doi: 10.3390/molecules27206831. Molecules. 2022. PMID: 36296424 Free PMC article.
-
Conditional Activation of Protein Therapeutics by Templated Removal of Peptide Nucleic Acid Masking Groups.Angew Chem Int Ed Engl. 2025 May;64(21):e202502268. doi: 10.1002/anie.202502268. Epub 2025 Mar 17. Angew Chem Int Ed Engl. 2025. PMID: 40062585 Free PMC article.
-
A fluorescent target-guided Paal-Knorr reaction.RSC Adv. 2020;10(61):37035-37039. doi: 10.1039/d0ra06962k. Epub 2020 Oct 7. RSC Adv. 2020. PMID: 34262697 Free PMC article.
-
Visible Light Photochemical Reactions for Nucleic Acid-Based Technologies.Molecules. 2021 Jan 21;26(3):556. doi: 10.3390/molecules26030556. Molecules. 2021. PMID: 33494512 Free PMC article. Review.
-
Selective Activation of Peptide-Thioester Precursors for Templated Native Chemical Ligations.Angew Chem Int Ed Engl. 2025 Jan 2;64(1):e202413644. doi: 10.1002/anie.202413644. Epub 2024 Oct 25. Angew Chem Int Ed Engl. 2025. PMID: 39198217 Free PMC article.
References
-
- Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1(1):13‐21. - PubMed
-
- Johnsson N, Johnsson K. Chemical tools for biomolecular imaging. ACS Chem Biol. 2007;2(1):31‐38. - PubMed
-
- Holstein JM, Rentmeister A. Current covalent modification methods for detecting RNA in fixed and living cells. Methods. 2016;98:18‐25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
