Expression of different ParE toxins results in conserved phenotypes with distinguishable classes of toxicity
- PMID: 31309747
- PMCID: PMC6813445
- DOI: 10.1002/mbo3.902
Expression of different ParE toxins results in conserved phenotypes with distinguishable classes of toxicity
Abstract
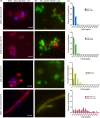
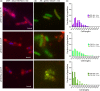
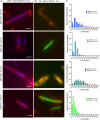
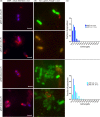
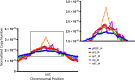
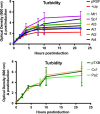
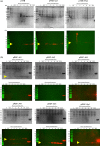
Toxin-antitoxin (TA) systems are found on both chromosomes and plasmids. These systems are unique in that they can confer both fatal and protective effects on bacterial cells-a quality that could potentially be harnessed given further understanding of these TA mechanisms. The current work focuses on the ParE subfamily, which is found throughout proteobacteria and has a sequence identity on average of approximately 12% (similarity at 30%-80%). Our aim is to evaluate the equivalency of chromosomally derived ParE toxin activity depending on its bacterial species of origin. Nine ParE toxins were analyzed, originating from six different bacterial species. Based on the resulting toxicity, three categories can be established: ParE toxins that do not exert toxicity under the experimental conditions, toxins that exert toxicity within the first four hours, and those that exert toxicity only after 10-12 hr of exposure. All tested ParE toxins produce a cellular morphologic change from rods to filaments, consistent with disruption of DNA topology. Analysis of the distribution of filamented cells within a population reveals a correlation between the extent of filamentation and toxicity. No membrane septation is visible along the length of the cell filaments, whereas aberrant lipid blebs are evident. Potent ParE-mediated toxicity is also correlated with a hallmark signature of abortive DNA replication, consistent with the inhibition of DNA gyrase.
Keywords: DNA gyrase; DNA topology; ParE toxin; filamented morphology; toxin-antitoxin systems.
© 2019 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Broken beyond repair: TA system ParE toxins mediate effective gyrase inhibition without driving resistance.J Bacteriol. 2025 Mar 20;207(3):e0041624. doi: 10.1128/jb.00416-24. Epub 2025 Mar 3. J Bacteriol. 2025. PMID: 40029095 Free PMC article.
-
Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin-antitoxin systems.Mol Microbiol. 2010 Jul 1;77(1):236-51. doi: 10.1111/j.1365-2958.2010.07207.x. Epub 2010 May 12. Mol Microbiol. 2010. PMID: 20487277 Free PMC article.
-
The three vibrio cholerae chromosome II-encoded ParE toxins degrade chromosome I following loss of chromosome II.J Bacteriol. 2011 Feb;193(3):611-9. doi: 10.1128/JB.01185-10. Epub 2010 Nov 29. J Bacteriol. 2011. PMID: 21115657 Free PMC article.
-
Ribonucleases in bacterial toxin-antitoxin systems.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):523-31. doi: 10.1016/j.bbagrm.2013.02.007. Epub 2013 Feb 21. Biochim Biophys Acta. 2013. PMID: 23454553 Review.
-
tRNAs taking charge.Pathog Dis. 2016 Mar;74(2):ftv117. doi: 10.1093/femspd/ftv117. Epub 2015 Dec 10. Pathog Dis. 2016. PMID: 26657107 Free PMC article. Review.
Cited by
-
Identifying a Molecular Mechanism That Imparts Species-Specific Toxicity to YoeB Toxins.Front Microbiol. 2020 May 21;11:959. doi: 10.3389/fmicb.2020.00959. eCollection 2020. Front Microbiol. 2020. PMID: 32528435 Free PMC article.
-
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.Sci Adv. 2024 Jan 5;10(1):eadj2403. doi: 10.1126/sciadv.adj2403. Epub 2024 Jan 5. Sci Adv. 2024. PMID: 38181072 Free PMC article.
-
Broken beyond repair: TA system ParE toxins mediate effective gyrase inhibition without driving resistance.J Bacteriol. 2025 Mar 20;207(3):e0041624. doi: 10.1128/jb.00416-24. Epub 2025 Mar 3. J Bacteriol. 2025. PMID: 40029095 Free PMC article.
-
Biology and evolution of bacterial toxin-antitoxin systems.Nat Rev Microbiol. 2022 Jun;20(6):335-350. doi: 10.1038/s41579-021-00661-1. Epub 2022 Jan 2. Nat Rev Microbiol. 2022. PMID: 34975154 Review.
-
Synsor: a tool for alignment-free detection of engineered DNA sequences.Front Bioeng Biotechnol. 2024 Jul 12;12:1375626. doi: 10.3389/fbioe.2024.1375626. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39070163 Free PMC article.
References
-
- Andersen, S. B. , Ghoul, M. , Griffin, A. S. , Petersen, B. , Johansen, H. K. , & Molin, S. (2017). Diversity, prevalence, and longitudinal occurrence of type II toxin‐antitoxin systems of Pseudomonas aeruginosa infecting cystic fibrosis lungs. Frontiers in Microbiology, 8, 1180 10.3389/fmicb.2017.01180 - DOI - PMC - PubMed
-
- Biasini, M. , Bienert, S. , Waterhouse, A. , Arnold, K. , Studer, G. , Schmidt, T. , … Schwede, T. (2014). SWISS‐MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 42(Web Server issue), W252–W258. 10.1093/nar/gku340 - DOI - PMC - PubMed
-
- Bos, J. , Zhang, Q. , Vyawahare, S. , Rogers, E. , Rosenberg, S. M. , & Austin, R. H. (2015). Emergence of antibiotic resistance from multinucleated bacterial filaments. Proceedings of the National Academy of Sciences of the United States of America, 112(1), 178–183. 10.1073/pnas.1420702111 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

