High-Resolution ENDOR Spectroscopy Combined with Quantum Chemical Calculations Reveals the Structure of Nitrogenase Janus Intermediate E4(4H)
- PMID: 31310109
- PMCID: PMC6956989
- DOI: 10.1021/jacs.9b04474
High-Resolution ENDOR Spectroscopy Combined with Quantum Chemical Calculations Reveals the Structure of Nitrogenase Janus Intermediate E4(4H)
Erratum in
-
Correction to "High-Resolution ENDOR Spectroscopy Combined with Quantum Chemical Calculations Reveals the Structure of Nitrogenase Janus Intermediate E4(4H)".J Am Chem Soc. 2019 Dec 18;141(50):19950. doi: 10.1021/jacs.9b13035. Epub 2019 Dec 10. J Am Chem Soc. 2019. PMID: 31820971 No abstract available.
Abstract
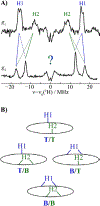
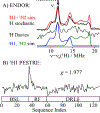
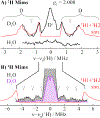
We have shown that the key state in N2 reduction to two NH3 molecules by the enzyme nitrogenase is E4(4H), the "Janus" intermediate, which has accumulated four [e-/H+] and is poised to undergo reductive elimination of H2 coupled to N2 binding and activation. Initial 1H and 95Mo ENDOR studies of freeze-trapped E4(4H) revealed that the catalytic multimetallic cluster (FeMo-co) binds two Fe-bridging hydrides, [Fe-H-Fe]. However, the analysis failed to provide a satisfactory picture of the relative spatial relationships of the two [Fe-H-Fe]. Our recent density functional theory (DFT) study yielded a lowest-energy form, denoted as E4(4H)(a), with two parallel Fe-H-Fe planes bridging pairs of "anchor" Fe on the Fe2,3,6,7 face of FeMo-co. However, the relative energies of structures E4(4H)(b), with one bridging and one terminal hydride, and E4(4H)(c), with one pair of anchor Fe supporting two bridging hydrides, were not beyond the uncertainties in the calculation. Moreover, a structure of V-dependent nitrogenase resulted in a proposed structure analogous to E4(4H)(c), and additional structures have been proposed in the DFT studies of others. To resolve the nature of hydride binding to the Janus intermediate, we performed exhaustive, high-resolution CW-stochastic 1H-ENDOR experiments using improved instrumentation, Mims 2H ENDOR, and a recently developed pulsed-ENDOR protocol ("PESTRE") to obtain absolute hyperfine interaction signs. These measurements are coupled to DFT structural models through an analytical point-dipole Hamiltonian for the hydride electron-nuclear dipolar coupling to its "anchoring" Fe ions, an approach that overcomes limitations inherent in both experimental interpretation and computational accuracy. The result is the freeze-trapped, lowest-energy Janus intermediate structure, E4(4H)(a).
Figures

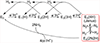







References
-
- Burgess BK; Lowe DJ Mechanism of Molybdenum Nitrogenase. Chem Rev 1996, 96, 2983–3012. - PubMed
-
- Thorneley RNF; Lowe DJ Kinetics and Mechanism of the Nitrogenase Enzyme System. Metal Ions in Biology 1985, 7, 221–284.
-
- Wilson PE; Nyborg AC; Watt GD Duplication and Extension of the Thorneley and Lowe Kinetic Model for Klebsiella Pneumoniae Nitrogenase Catalysis Using a Mathematica Software Platform. Biophysical Chemistry 2001, 91, 281–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

