Human indole(ethyl)amine-N-methyltransferase (hINMT) catalyzed methylation of tryptamine, dimethylsulfide and dimethylselenide is enhanced under reducing conditions - A comparison between 254C and 254F, two common hINMT variants
- PMID: 31310642
- PMCID: PMC6634407
- DOI: 10.1371/journal.pone.0219664
Human indole(ethyl)amine-N-methyltransferase (hINMT) catalyzed methylation of tryptamine, dimethylsulfide and dimethylselenide is enhanced under reducing conditions - A comparison between 254C and 254F, two common hINMT variants
Erratum in
-
Correction: Human indole(ethyl)amine-N-methyltransferase (hINMT) catalyzed methylation of tryptamine, dimethylsulfide and dimethylselenide is enhanced under reducing conditions - A comparison between 254C and 254F, two common hINMT variants.PLoS One. 2019 Oct 1;14(10):e0223546. doi: 10.1371/journal.pone.0223546. eCollection 2019. PLoS One. 2019. PMID: 31574131 Free PMC article.
Abstract
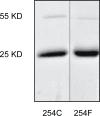
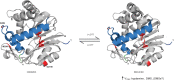
Phenylalanine and cysteine comprise common miss-sense variants (i.e., single nucleotide polymorphisms [SNPs]) at amino acid position 254 of the human indole(ethyl)amine-N-methyltransferase (hINMT). The phenylalanine variant, which occurs in linkage disequilibrium with two 3' UTR SNPs, has been reported to associate with elevated urine levels of trimethylselenonium (TMSe), the Se-methylated product of volatile dimethylselenide. hINMT allozymes expressing either cysteine (254C) or phenylalanine (254F) at position 254 were compared for enzyme activity (i.e., Km and Vmax) towards the INMT substrates tryptamine, dimethylsulfide (DMS) and dimethylselenide (DMSe) in vitro. The SNP 254C had a higher Vmax for DMS and tryptamine in the presence of reducing agent than in its absence. Conversely, Vmax for 254F was insensitive to the presence or absence of reducing agent for these substrates. SNP 254F showed a lower Km for tryptamine in the absence of reducing agent than 254C. No statistically significant difference in Vmax or Km was observed between 254C and 254F allozymes in the presence of reducing agent for DMSe, The Km values for DMSe methylation were about 10-fold (254C) or 6-fold (254F) more favorable than for tryptamine methylation with reducing agent present. These findings indicated that: 1) That phenylalanine at position 254 renders hINMT methylation of substrates DMS and tryptamine insensitive to a non reducing environment. 2) That human INMT harbors significant thioether-S-methyltransferase (TEMT) activity with a higher affinity for DMSe than tryptamine, 3) The reduction of a 44C/254C disulfide bond in hINMT that increases Vmax is proposed.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

