Convergent evolution of cytochrome P450s underlies independent origins of keto-carotenoid pigmentation in animals
- PMID: 31311468
- PMCID: PMC6661338
- DOI: 10.1098/rspb.2019.1039
Convergent evolution of cytochrome P450s underlies independent origins of keto-carotenoid pigmentation in animals
Abstract
Keto-carotenoids contribute to many important traits in animals, including vision and coloration. In a great number of animal species, keto-carotenoids are endogenously produced from carotenoids by carotenoid ketolases. Despite the ubiquity and functional importance of keto-carotenoids in animals, the underlying genetic architectures of their production have remained enigmatic. The body and eye colorations of spider mites (Arthropoda: Chelicerata) are determined by β-carotene and keto-carotenoid derivatives. Here, we focus on a carotenoid pigment mutant of the spider mite Tetranychus kanzawai that, as shown by chromatography, lost the ability to produce keto-carotenoids. We employed bulked segregant analysis and linked the causal locus to a single narrow genomic interval. The causal mutation was fine-mapped to a minimal candidate region that held only one complete gene, the cytochrome P450 monooxygenase CYP384A1, of the CYP3 clan. Using a number of genomic approaches, we revealed that an inactivating deletion in the fourth exon of CYP384A1 caused the aberrant pigmentation. Phylogenetic analysis indicated that CYP384A1 is orthologous across mite species of the ancient Trombidiformes order where carotenoids typify eye and body coloration, suggesting a deeply conserved function of CYP384A1 as a carotenoid ketolase. Previously, CYP2J19, a cytochrome P450 of the CYP2 clan, has been identified as a carotenoid ketolase in birds and turtles. Our study shows that selection for endogenous production of keto-carotenoids led to convergent evolution, whereby cytochrome P450s were independently co-opted in vertebrate and invertebrate animal lineages.
Keywords: CYP384A1; carotenoid ketolase; convergent evolution; keto-carotenoids; lemon.
Conflict of interest statement
We declare we have no competing interests.
Figures


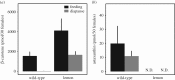
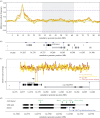
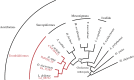
References
-
- Goodwin TW. 1984. The biochemistry of the carotenoids, volume II: animals. London, UK: Chapman and Hall.
-
- Hill GE, McGraw KJ. 2006. Bird coloration, vol. 2: function and evolution. Cambridge, MA: Harvard University Press.
-
- Bosse TC, Veerman A. 1996. Involvement of vitamin A in the photoperiodic induction of diapause in the spider mite Tetranychus urticae is demonstrated by rearing an albino mutant on a semi-synthetic diet with and without p-carotene or vitamin A. Physiol. Entomol. 21, 188–192. (10.1111/j.1365-3032.1996.tb00854.x) - DOI
-
- Heath JJ, Cipollini DF, Stireman JO III. 2013. The role of carotenoids and their derivatives in mediating interactions between insects and their environment. Arthropod-Plant Interact. 7, 1–20. (10.1007/s11829-012-9239-7) - DOI
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
