Application of a MYC degradation screen identifies sensitivity to CDK9 inhibitors in KRAS-mutant pancreatic cancer
- PMID: 31311847
- PMCID: PMC6728149
- DOI: 10.1126/scisignal.aav7259
Application of a MYC degradation screen identifies sensitivity to CDK9 inhibitors in KRAS-mutant pancreatic cancer
Abstract
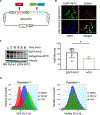
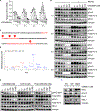
Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adenocarcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry-based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage-dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser62 Our study thus not only identifies a potential therapeutic target for patients with KRAS-mutant PDAC but also presents the application of a screening strategy that can be more broadly adapted to identify regulators of protein stability.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures




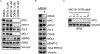
References
-
- Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018. CA Cancer J Clin 68, 7–30 (2018). - PubMed
-
- Rahib L et al., Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74, 2913–2921 (2014). - PubMed
-
- Ryan DP, Hong TS, Bardeesy N, Pancreatic adenocarcinoma. N Engl J Med 371, 2140–2141 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

