Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates
- PMID: 31311879
- PMCID: PMC6635527
- DOI: 10.1128/mBio.01083-19
Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates
Abstract
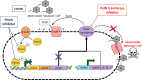
Nosocomial infections with Acinetobacter baumannii are a global problem in intensive care units with high mortality rates. Increasing resistance to first- and second-line antibiotics has forced the use of colistin as last-resort treatment, and increasing development of colistin resistance in A. baumannii has been reported. We evaluated the transcriptional regulator PmrA as potential drug target to restore colistin efficacy in A. baumannii Deletion of pmrA restored colistin susceptibility in 10 of the 12 extensively drug-resistant A. baumannii clinical isolates studied, indicating the importance of PmrA in the drug resistance phenotype. However, two strains remained highly resistant, indicating that PmrA-mediated overexpression of the phosphoethanolamine (PetN) transferase PmrC is not the exclusive colistin resistance mechanism in A. baumannii A detailed genetic characterization revealed a new colistin resistance mechanism mediated by genetic integration of the insertion element ISAbaI upstream of the PmrC homolog EptA (93% identity), leading to its overexpression. We found that eptA was ubiquitously present in clinical strains belonging to the international clone 2, and ISAbaI integration upstream of eptA was required to mediate the colistin-resistant phenotype. In addition, we found a duplicated ISAbaI-eptA cassette in one isolate, indicating that this colistin resistance determinant may be embedded in a mobile genetic element. Our data disprove PmrA as a drug target for adjuvant therapy but highlight the importance of PetN transferase-mediated colistin resistance in clinical strains. We suggest that direct targeting of the homologous PetN transferases PmrC/EptA may have the potential to overcome colistin resistance in A. baumanniiIMPORTANCE The discovery of antibiotics revolutionized modern medicine and enabled us to cure previously deadly bacterial infections. However, a progressive increase in antibiotic resistance rates is a major and global threat for our health care system. Colistin represents one of our last-resort antibiotics that is still active against most Gram-negative bacterial pathogens, but increasing resistance is reported worldwide, in particular due to the plasmid-encoded protein MCR-1 present in pathogens such as Escherichia coli and Klebsiella pneumoniae Here, we showed that colistin resistance in A. baumannii, a top-priority pathogen causing deadly nosocomial infections, is mediated through different avenues that result in increased activity of homologous phosphoethanolamine (PetN) transferases. Considering that MCR-1 is also a PetN transferase, our findings indicate that PetN transferases might be the Achilles heel of superbugs and that direct targeting of them may have the potential to preserve the activity of polymyxin antibiotics.
Keywords: Acinetobacter baumannii; antibiotic resistance; colistin; eptA; ethanolamine transferase; mcr-1; pmrA.
Copyright © 2019 Trebosc et al.
Figures


References
-
- Boucher HW, Talbot GH, Benjamin DK, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Diseases Society of America. 2013. 10 × ’20 progress—development of new drugs active against gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi:10.1093/cid/cit152. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
