Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients
- PMID: 31311881
- PMCID: PMC6635529
- DOI: 10.1128/mBio.01315-19
Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients
Abstract
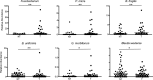
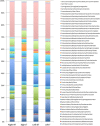
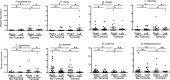
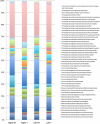
We have previously identified a crypt-specific core microbiota (CSCM) in the colons of healthy laboratory mice and related wild rodents. Here, we confirm that a CSCM also exists in the human colon and appears to be altered during colon cancer. The colonic microbiota is suggested to be involved in the development of colorectal cancer (CRC). Because the microbiota identified in fecal samples from CRC patients does not directly reflect the microbiota associated with tumor tissues themselves, we sought to characterize the bacterial communities from the crypts and associated adjacent mucosal surfaces of 58 patients (tumor and normal homologous tissue) and 9 controls with normal colonoscopy results. Here, we confirm that bacteria colonize human colonic crypts in both control and CRC tissues, and using laser-microdissected tissues and 16S rRNA gene sequencing, we further show that right and left crypt- and mucosa-associated bacterial communities are significantly different. In addition to Bacteroidetes and Firmicutes, and as with murine proximal colon crypts, environmental nonfermentative Proteobacteria are found in human colonic crypts. Fusobacterium and Bacteroides fragilis are more abundant in right-side tumors, whereas Parvimonas micra is more prevalent in left-side tumors. More precisely, Fusobacterium periodonticum is more abundant in crypts from cancerous samples in the right colon than in associated nontumoral samples from adjacent areas but not in left-side colonic samples. Future analysis of the interaction between these bacteria and the crypt epithelium, particularly intestinal stem cells, will allow deciphering of their possible oncogenic potential.IMPORTANCE Due to the huge number of bacteria constituting the human colon microbiota, alteration in the balance of its constitutive taxa (i.e., dysbiosis) is highly suspected of being involved in colorectal oncogenesis. Indeed, bacterial signatures in association with CRC have been described. These signatures may vary if bacteria are identified in feces or in association with tumor tissues. Here, we show that bacteria colonize human colonic crypts in tissues obtained from patients with CRC and with normal colonoscopy results. Aerobic nonfermentative Proteobacteria previously identified as constitutive of the crypt-specific core microbiota in murine colonic samples are similarly prevalent in human colonic crypts in combination with other anaerobic taxa. We also show that bacterial signatures characterizing the crypts of colonic tumors vary depending whether right-side or left-side tumors are analyzed.
Keywords: colon cancer; intestinal crypts; microbiota.
Copyright © 2019 Saffarian et al.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
