The genomic landscape of Epstein-Barr virus-associated pulmonary lymphoepithelioma-like carcinoma
- PMID: 31311932
- PMCID: PMC6635366
- DOI: 10.1038/s41467-019-10902-w
The genomic landscape of Epstein-Barr virus-associated pulmonary lymphoepithelioma-like carcinoma
Abstract
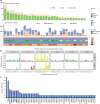
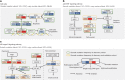
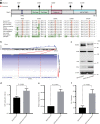
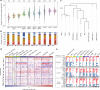
Pulmonary lymphoepithelioma-like carcinoma (LELC) is a rare and distinct subtype of primary lung cancer characterized by Epstein-Barr virus (EBV) infection. Herein, we reported the mutational landscape of pulmonary LELC using whole-exome sequencing, targeted deep sequencing and single-nucleotide polymorphism arrays. We identify a low degree of somatic mutation but widespread existence of copy number variations. We reveal predominant signature 2 mutations and frequent loss of type I interferon genes that are involved in the host-virus counteraction. Integrated analysis shows enrichment of genetic lesions affecting several critical pathways, including NF-κB, JAK/STAT, and cell cycle. Notably, multi-dimensional comparison unveils that pulmonary LELC resemble NPC but are clearly different from other lung cancers, natural killer/T-cell lymphoma or EBV-related gastric cancer in terms of genetic features. In all, our study illustrates a distinct genomic landscape of pulmonary LELC and provides a road map to facilitate genome-guided personalized treatment.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

